Three-dimensional chromatin interactions remain stable upon CAG/CTG repeat expansion
- PMID: 32656337
- PMCID: PMC7334000
- DOI: 10.1126/sciadv.aaz4012
Three-dimensional chromatin interactions remain stable upon CAG/CTG repeat expansion
Abstract
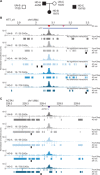
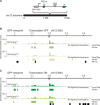
Expanded CAG/CTG repeats underlie 13 neurological disorders, including myotonic dystrophy type 1 (DM1) and Huntington's disease (HD). Upon expansion, disease loci acquire heterochromatic characteristics, which may provoke changes to chromatin conformation and thereby affect both gene expression and repeat instability. Here, we tested this hypothesis by performing 4C sequencing at the DMPK and HTT loci from DM1 and HD-derived cells. We find that allele sizes ranging from 15 to 1700 repeats displayed similar chromatin interaction profiles. This was true for both loci and for alleles with different DNA methylation levels and CTCF binding. Moreover, the ectopic insertion of an expanded CAG repeat tract did not change the conformation of the surrounding chromatin. We conclude that CAG/CTG repeat expansions are not enough to alter chromatin conformation in cis. Therefore, it is unlikely that changes in chromatin interactions drive repeat instability or changes in gene expression in these disorders.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures


References
-
- Bickmore W. A., van Steensel B., Genome architecture: Domain organization of interphase chromosomes. Cell 152, 1270–1284 (2013). - PubMed
-
- Kempfer R., Pombo A., Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 21, 207–226 (2020). - PubMed
-
- Ibrahim D. M., Mundlos S., Three-dimensional chromatin in disease: What holds us together and what drives us apart? Curr. Opin. Cell Biol. 64, 1–9 (2020). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

