L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer
- PMID: 32656539
- PMCID: PMC7351134
- DOI: 10.1038/s43018-019-0006-x
L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer
Erratum in
-
Author Correction: L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer.Nat Cancer. 2020 Nov;1(11):1128. doi: 10.1038/s43018-020-00130-3. Nat Cancer. 2020. PMID: 35122071 No abstract available.
Abstract
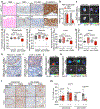
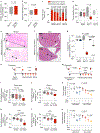
Metastasis-initiating cells with stem-like properties drive cancer lethality, yet their origins and relationship to primary-tumor-initiating stem cells are not known. We show that L1CAM+ cells in human colorectal cancer (CRC) have metastasis-initiating capacity, and we define their relationship to tissue regeneration. L1CAM is not expressed in the homeostatic intestinal epithelium, but is induced and required for epithelial regeneration following colitis and in CRC organoid growth. By using human tissues and mouse models, we show that L1CAM is dispensable for adenoma initiation but required for orthotopic carcinoma propagation, liver metastatic colonization and chemoresistance. L1CAMhigh cells partially overlap with LGR5high stem-like cells in human CRC organoids. Disruption of intercellular epithelial contacts causes E-cadherin-REST transcriptional derepression of L1CAM, switching chemoresistant CRC progenitors from an L1CAMlow to an L1CAMhigh state. Thus, L1CAM dependency emerges in regenerative intestinal cells when epithelial integrity is lost, a phenotype of wound healing deployed in metastasis-initiating cells.
Conflict of interest statement
Competing interests J.M. is a science advisor for and owns company stock in Scholar Rock.
Figures












Comment in
-
L1CAM links regeneration to metastasis.Nat Cancer. 2020 Jan;1(1):22-24. doi: 10.1038/s43018-019-0014-x. Nat Cancer. 2020. PMID: 35121841 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

