Harnessing endophenotypes and network medicine for Alzheimer's drug repurposing
- PMID: 32656864
- PMCID: PMC7561446
- DOI: 10.1002/med.21709
Harnessing endophenotypes and network medicine for Alzheimer's drug repurposing
Abstract
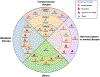
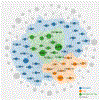
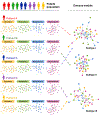
Following two decades of more than 400 clinical trials centered on the "one drug, one target, one disease" paradigm, there is still no effective disease-modifying therapy for Alzheimer's disease (AD). The inherent complexity of AD may challenge this reductionist strategy. Recent observations and advances in network medicine further indicate that AD likely shares common underlying mechanisms and intermediate pathophenotypes, or endophenotypes, with other diseases. In this review, we consider AD pathobiology, disease comorbidity, pleiotropy, and therapeutic development, and construct relevant endophenotype networks to guide future therapeutic development. Specifically, we discuss six main endophenotype hypotheses in AD: amyloidosis, tauopathy, neuroinflammation, mitochondrial dysfunction, vascular dysfunction, and lysosomal dysfunction. We further consider how this endophenotype network framework can provide advances in computational and experimental strategies for drug-repurposing and identification of new candidate therapeutic strategies for patients suffering from or at risk for AD. We highlight new opportunities for endophenotype-informed, drug discovery in AD, by exploiting multi-omics data. Integration of genomics, transcriptomics, radiomics, pharmacogenomics, and interactomics (protein-protein interactions) are essential for successful drug discovery. We describe experimental technologies for AD drug discovery including human induced pluripotent stem cells, transgenic mouse/rat models, and population-based retrospective case-control studies that may be integrated with multi-omics in a network medicine methodology. In summary, endophenotype-based network medicine methodologies will promote AD therapeutic development that will optimize the usefulness of available data and support deep phenotyping of the patient heterogeneity for personalized medicine in AD.
Keywords: Alzheimer's disease; amyloidosis; drug repurposing; endophenotype; network medicine; omics; pathobiology; systems biology; tauopathy.
© 2020 Wiley Periodicals LLC.
Figures






References
-
- Alzheimer A Uber eine eigenartige Erkrankung der Hirnrinde. Zentralbl Nervenh Psych 1907;18:177–179.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

