Controlling gene expression with light: a multidisciplinary endeavour
- PMID: 32657338
- PMCID: PMC7458398
- DOI: 10.1042/BST20200014
Controlling gene expression with light: a multidisciplinary endeavour
Abstract
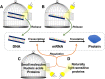
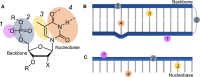
The expression of a gene to a protein is one of the most vital biological processes. The use of light to control biology offers unparalleled spatiotemporal resolution from an external, orthogonal signal. A variety of methods have been developed that use light to control the steps of transcription and translation of specific genes into proteins, for cell-free to in vivo biotechnology applications. These methods employ techniques ranging from the modification of small molecules, nucleic acids and proteins with photocages, to the engineering of proteins involved in gene expression using naturally light-sensitive proteins. Although the majority of currently available technologies employ ultraviolet light, there has been a recent increase in the use of functionalities that work at longer wavelengths of light, to minimise cellular damage and increase tissue penetration. Here, we discuss the different chemical and biological methods employed to control gene expression, while also highlighting the central themes and the most exciting applications within this diverse field.
Keywords: biochemical techniques and resources; biotechnology; gene expression and regulation.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

