Distance indexing and seed clustering in sequence graphs
- PMID: 32657356
- PMCID: PMC7355256
- DOI: 10.1093/bioinformatics/btaa446
Distance indexing and seed clustering in sequence graphs
Abstract
Motivation: Graph representations of genomes are capable of expressing more genetic variation and can therefore better represent a population than standard linear genomes. However, due to the greater complexity of genome graphs relative to linear genomes, some functions that are trivial on linear genomes become much more difficult in genome graphs. Calculating distance is one such function that is simple in a linear genome but complicated in a graph context. In read mapping algorithms such distance calculations are fundamental to determining if seed alignments could belong to the same mapping.
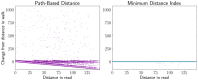
Results: We have developed an algorithm for quickly calculating the minimum distance between positions on a sequence graph using a minimum distance index. We have also developed an algorithm that uses the distance index to cluster seeds on a graph. We demonstrate that our implementations of these algorithms are efficient and practical to use for a new generation of mapping algorithms based upon genome graphs.
Availability and implementation: Our algorithms have been implemented as part of the vg toolkit and are available at https://github.com/vgteam/vg.
© The Author(s) 2020. Published by Oxford University Press.
Figures








References
-
- Akiba T. et al. (2013) Fast exact shortest-path distance queries on large networks by pruned landmark labeling. In Proceedings of the 2013 International Conference on Management of Data - SIGMOD’13, ACM Press, New York, NY, USA, p. 349.
-
- Dave V.S., Hasan M.A. (2015) TopCom: index for shortest distance query in directed graph In Q Chen. et al. (eds) Database and Expert Systems Applications, Lecture Notes in Computer Science, Springer International Publishing, Cham, pp. 471–480.
-
- Dijkstra E.W. (1959) A note on two problems in connexion with graphs. Numer. Math., 1, 269–271.
-
- Djidjev H.N. (1997) Efficient algorithms for shortest path queries in planar digraphs In: G. Gooset al. (eds) Graph-Theoretic Concepts in Computer Science. Vol. 1197, Springer, Berlin, Heidelberg, pp. 151–165.

