Weighted minimizer sampling improves long read mapping
- PMID: 32657365
- PMCID: PMC7355284
- DOI: 10.1093/bioinformatics/btaa435
Weighted minimizer sampling improves long read mapping
Abstract
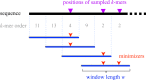
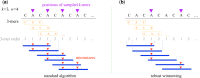
Motivation: In this era of exponential data growth, minimizer sampling has become a standard algorithmic technique for rapid genome sequence comparison. This technique yields a sub-linear representation of sequences, enabling their comparison in reduced space and time. A key property of the minimizer technique is that if two sequences share a substring of a specified length, then they can be guaranteed to have a matching minimizer. However, because the k-mer distribution in eukaryotic genomes is highly uneven, minimizer-based tools (e.g. Minimap2, Mashmap) opt to discard the most frequently occurring minimizers from the genome to avoid excessive false positives. By doing so, the underlying guarantee is lost and accuracy is reduced in repetitive genomic regions.
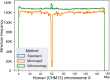
Results: We introduce a novel weighted-minimizer sampling algorithm. A unique feature of the proposed algorithm is that it performs minimizer sampling while considering a weight for each k-mer; i.e. the higher the weight of a k-mer, the more likely it is to be selected. By down-weighting frequently occurring k-mers, we are able to meet both objectives: (i) avoid excessive false-positive matches and (ii) maintain the minimizer match guarantee. We tested our algorithm, Winnowmap, using both simulated and real long-read data and compared it to a state-of-the-art long read mapper, Minimap2. Our results demonstrate a reduction in the mapping error-rate from 0.14% to 0.06% in the recently finished human X chromosome (154.3 Mbp), and from 3.6% to 0% within the highly repetitive X centromere (3.1 Mbp). Winnowmap improves mapping accuracy within repeats and achieves these results with sparser sampling, leading to better index compression and competitive runtimes.
Availability and implementation: Winnowmap is built on top of the Minimap2 codebase and is available at https://github.com/marbl/winnowmap.
Published by Oxford University Press 2020.
Figures


References
-
- Berlin K. et al. (2015) Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol., 33, 623–630. - PubMed
-
- Broder A.Z. (1997) On the resemblance and containment of documents. In Proceedings Compression and Complexity of Sequences 1997 (Cat. No. 97TB100171), pp. 21–29. IEEE.
-
- Chikhi R. et al. (2015) On the representation of de Bruijn graphs. J. Comput. Biol., 22, 336–352. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

