Proteomics and Informatics for Understanding Phases and Identifying Biomarkers in COVID-19 Disease
- PMID: 32657586
- PMCID: PMC7384384
- DOI: 10.1021/acs.jproteome.0c00326
Proteomics and Informatics for Understanding Phases and Identifying Biomarkers in COVID-19 Disease
Abstract
The emergence of novel coronavirus disease 2019 (COVID-19), caused by the SARS-CoV-2 coronavirus, has necessitated the urgent development of new diagnostic and therapeutic strategies. Rapid research and development, on an international scale, has already generated assays for detecting SARS-CoV-2 RNA and host immunoglobulins. However, the complexities of COVID-19 are such that fuller definitions of patient status, trajectory, sequelae, and responses to therapy are now required. There is accumulating evidence-from studies of both COVID-19 and the related disease SARS-that protein biomarkers could help to provide this definition. Proteins associated with blood coagulation (D-dimer), cell damage (lactate dehydrogenase), and the inflammatory response (e.g., C-reactive protein) have already been identified as possible predictors of COVID-19 severity or mortality. Proteomics technologies, with their ability to detect many proteins per analysis, have begun to extend these early findings. To be effective, proteomics strategies must include not only methods for comprehensive data acquisition (e.g., using mass spectrometry) but also informatics approaches via which to derive actionable information from large data sets. Here we review applications of proteomics to COVID-19 and SARS and outline how pipelines involving technologies such as artificial intelligence could be of value for research on these diseases.
Keywords: COVID-19; SARS-CoV-2; artificial intelligence; assay; biomarker; diagnosis; marker; prognosis; proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
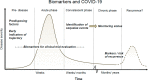


References
-
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395 (10223), 497–506. 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Li Q.; Guan X.; Wu P.; Wang X.; Zhou L.; Tong Y.; Ren R.; Leung K. S. M.; Lau E. H. Y.; Wong J. Y.; Xing X.; Xiang N.; Wu Y.; Li C.; Chen Q.; Li D.; Liu T.; Zhao J.; Liu M.; Tu W.; Chen C.; Jin L.; Yang R.; Wang Q.; Zhou S.; Wang R.; Liu H.; Luo Y.; Liu Y.; Shao G.; Li H.; Tao Z.; Yang Y.; Deng Z.; Liu B.; Ma Z.; Zhang Y.; Shi G.; Lam T. T. Y.; Wu J. T.; Gao G. F.; Cowling B. J.; Yang B.; Leung G. M.; Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382 (13), 1199–1207. 10.1056/NEJMoa2001316. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

