Large-scale assessment of antimicrobial resistance marker databases for genetic phenotype prediction: a systematic review
- PMID: 32658975
- PMCID: PMC7566382
- DOI: 10.1093/jac/dkaa257
Large-scale assessment of antimicrobial resistance marker databases for genetic phenotype prediction: a systematic review
Abstract
Background: Antimicrobial resistance (AMR) is a rising health threat with 10 million annual casualties estimated by 2050. Appropriate treatment of infectious diseases with the right antibiotics reduces the spread of antibiotic resistance. Today, clinical practice relies on molecular and PCR techniques for pathogen identification and culture-based antibiotic susceptibility testing (AST). Recently, WGS has started to transform clinical microbiology, enabling prediction of resistance phenotypes from genotypes and allowing for more informed treatment decisions. WGS-based AST (WGS-AST) depends on the detection of AMR markers in sequenced isolates and therefore requires AMR reference databases. The completeness and quality of these databases are material to increase WGS-AST performance.
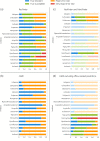
Methods: We present a systematic evaluation of the performance of publicly available AMR marker databases for resistance prediction on clinical isolates. We used the public databases CARD and ResFinder with a final dataset of 2587 isolates across five clinically relevant pathogens from PATRIC and NDARO, public repositories of antibiotic-resistant bacterial isolates.
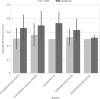
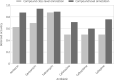
Results: CARD and ResFinder WGS-AST performance had an overall balanced accuracy of 0.52 (±0.12) and 0.66 (±0.18), respectively. Major error rates were higher in CARD (42.68%) than ResFinder (25.06%). However, CARD showed almost no very major errors (1.17%) compared with ResFinder (4.42%).
Conclusions: We show that AMR databases need further expansion, improved marker annotations per antibiotic rather than per antibiotic class and validated multivariate marker panels to achieve clinical utility, e.g. in order to meet performance requirements such as provided by the FDA for clinical microbiology diagnostic testing.
© The Author(s) 2020. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy.
Figures


References
-
- O’Neill J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. The Review on Antimicrobial Resistance 2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Ta....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

