RNA-Dependent RNA Polymerase as a Target for COVID-19 Drug Discovery
- PMID: 32660307
- PMCID: PMC7684788
- DOI: 10.1177/2472555220942123
RNA-Dependent RNA Polymerase as a Target for COVID-19 Drug Discovery
Abstract
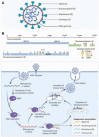
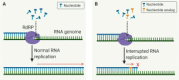
COVID-19 respiratory disease caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has rapidly become a global health issue since it emerged in December 2019. While great global efforts are underway to develop vaccines and to discover or repurpose therapeutic agents for this disease, as of this writing only the nucleoside drug remdesivir has been approved under Emergency Use Authorization to treat COVID-19. The RNA-dependent RNA polymerase (RdRP), a viral enzyme for viral RNA replication in host cells, is one of the most intriguing and promising drug targets for SARS-CoV-2 drug development. Because RdRP is a viral enzyme with no host cell homologs, selective SARS-CoV-2 RdRP inhibitors can be developed that have improved potency and fewer off-target effects against human host proteins and thus are safer and more effective therapeutics for treating COVID-19. This review focuses on biochemical enzyme and cell-based assays for RdRPs that could be used in high-throughput screening to discover new and repurposed drugs against SARS-CoV-2.
Keywords: COVID-19; RNA-dependent RNA polymerase; RdRP assays; RdRP inhibitors; SARS-CoV-2; coronavirus infection.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

