Bioinformatic analysis reveals MIR502 as a potential tumour suppressor in ovarian cancer
- PMID: 32660514
- PMCID: PMC7359466
- DOI: 10.1186/s13048-020-00683-y
Bioinformatic analysis reveals MIR502 as a potential tumour suppressor in ovarian cancer
Abstract
Background: Ovarian cancer (OC) is a major cause of death among women due to the lack of early screening methods and its complex pathological progression. Increasing evidence has indicated that microRNAs regulate gene expression in tumours by interacting with mRNAs. Although the research regarding OC and microRNAs is extensive, the vital role of MIR502 in OC remains unclear.
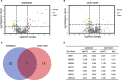
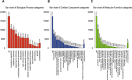
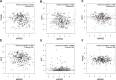
Methods: We integrated two microRNA expression arrays from GEO to identify differentially expressed genes. The Kaplan-Meier method was used to screen for miRNAs that had an influence on survival outcome. Upstream regulators of MIR502 were predicted by JASPAR and verified by ChIP-seq data. The LinkedOmics database was used to study genes that were correlated with MIR502. Gene Set Enrichment Analysis (GSEA) was conducted for functional annotation with GO and KEGG pathway enrichment analyses by using the open access WebGestalt tool. We constructed a PPI network by using STRING to further explore the core proteins.
Results: We found that the expression level of MIR502 was significantly downregulated in OC, which was related to poor overall survival. NRF1, as an upstream regulator of MIR502, was predicted by JASPAR and verified by ChIP-seq data. In addition, anti-apoptosis and pro-proliferation genes in the Hippo signalling pathway, including CCND1, MYC, FGF1 and GLI2, were negatively regulated by MIR502, as shown in the GO and KEGG pathway enrichment results. The PPI network further demonstrated that CCND1 and MYCN were at core positions in the development of ovarian cancer.
Conclusions: MIR502, which is regulated by NRF1, acts as a tumour suppressor gene to accelerate apoptosis and suppress proliferation by targeting the Hippo signalling pathway in ovarian cancer.
Keywords: Hippo signalling pathway; MIR502; NRF1; Ovarian cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- La Vecchia C. Ovarian cancer: epidemiology and risk factors. Eur J Cancer Prev. 2017;26:55–62. - PubMed
-
- Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram‐Baptiste D, Saslow D, et al. Cancer screening in the United States, 2018: a review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2018;68:297–316. - PubMed
-
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

