Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2
- PMID: 32661139
- PMCID: PMC7459545
- DOI: 10.1128/JVI.00940-20
Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2
Abstract
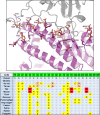
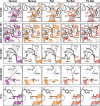
The COVID-19 pandemic has caused an unprecedented global public health and economic crisis. The origin and emergence of its causal agent, SARS-CoV-2, in the human population remains mysterious, although bat and pangolin were proposed to be the natural reservoirs. Strikingly, unlike the SARS-CoV-2-like coronaviruses (CoVs) identified in bats and pangolins, SARS-CoV-2 harbors a polybasic furin cleavage site in its spike (S) glycoprotein. SARS-CoV-2 uses human angiotensin-converting enzyme 2 (ACE2) as its receptor to infect cells. Receptor recognition by the S protein is the major determinant of host range, tissue tropism, and pathogenesis of coronaviruses. In an effort to search for the potential intermediate or amplifying animal hosts of SARS-CoV-2, we examined receptor activity of ACE2 from 14 mammal species and found that ACE2s from multiple species can support the infectious entry of lentiviral particles pseudotyped with the wild-type or furin cleavage site-deficient S protein of SARS-CoV-2. ACE2 of human/rhesus monkey and rat/mouse exhibited the highest and lowest receptor activities, respectively. Among the remaining species, ACE2s from rabbit and pangolin strongly bound to the S1 subunit of SARS-CoV-2 S protein and efficiently supported the pseudotyped virus infection. These findings have important implications for understanding potential natural reservoirs, zoonotic transmission, human-to-animal transmission, and use of animal models.IMPORTANCE SARS-CoV-2 uses human ACE2 as a primary receptor for host cell entry. Viral entry mediated by the interaction of ACE2 with spike protein largely determines host range and is the major constraint to interspecies transmission. We examined the receptor activity of 14 ACE2 orthologs and found that wild-type and mutant SARS-CoV-2 lacking the furin cleavage site in S protein could utilize ACE2 from a broad range of animal species to enter host cells. These results have important implications in the natural hosts, interspecies transmission, animal models, and molecular basis of receptor binding for SARS-CoV-2.
Keywords: SARS-CoV-2; animal ACE2; animal hosts; entry; furin cleavage; receptor.
Copyright © 2020 Zhao et al.
Figures
Similar articles
-
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article.
-
Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts.J Virol. 2020 Jul 16;94(15):e00831-20. doi: 10.1128/JVI.00831-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32404529 Free PMC article.
-
Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2.Proc Natl Acad Sci U S A. 2021 Mar 23;118(12):e2025373118. doi: 10.1073/pnas.2025373118. Proc Natl Acad Sci U S A. 2021. PMID: 33658332 Free PMC article.
-
SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy.Theranostics. 2020 Jun 12;10(16):7448-7464. doi: 10.7150/thno.48076. eCollection 2020. Theranostics. 2020. PMID: 32642005 Free PMC article. Review.
-
Angiotensin-converting enzyme 2 (ACE2) receptor and SARS-CoV-2: Potential therapeutic targeting.Eur J Pharmacol. 2020 Oct 5;884:173455. doi: 10.1016/j.ejphar.2020.173455. Epub 2020 Jul 31. Eur J Pharmacol. 2020. PMID: 32745604 Free PMC article. Review.
Cited by
-
Intrinsic host susceptibility among multiple species to intranasal SARS-CoV-2 identifies diverse virological, biodistribution and pathological outcomes.Sci Rep. 2022 Nov 4;12(1):18694. doi: 10.1038/s41598-022-23339-x. Sci Rep. 2022. PMID: 36333445 Free PMC article.
-
Animal models and SARS-CoV-2-induced pulmonary and neurological injuries.Mem Inst Oswaldo Cruz. 2023 Jan 20;117:e220239. doi: 10.1590/0074-02760220239. eCollection 2023. Mem Inst Oswaldo Cruz. 2023. PMID: 36700583 Free PMC article. Review.
-
Variations in Cell Surface ACE2 Levels Alter Direct Binding of SARS-CoV-2 Spike Protein and Viral Infectivity: Implications for Measuring Spike Protein Interactions with Animal ACE2 Orthologs.J Virol. 2022 Sep 14;96(17):e0025622. doi: 10.1128/jvi.00256-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000847 Free PMC article.
-
SARS-CoV-2 Infection: New Molecular, Phylogenetic, and Pathogenetic Insights. Efficacy of Current Vaccines and the Potential Risk of Variants.Viruses. 2021 Aug 25;13(9):1687. doi: 10.3390/v13091687. Viruses. 2021. PMID: 34578269 Free PMC article. Review.
-
Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.PLoS Pathog. 2023 Apr 5;19(4):e1011206. doi: 10.1371/journal.ppat.1011206. eCollection 2023 Apr. PLoS Pathog. 2023. PMID: 37018380 Free PMC article.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi:10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi:10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
-
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC, Bi Y, Shi W. 2020. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30:2196–2203.e2193. doi:10.1016/j.cub.2020.05.023. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

