Structural biology of DNA abasic site protection by SRAP proteins
- PMID: 32663791
- PMCID: PMC7494546
- DOI: 10.1016/j.dnarep.2020.102903
Structural biology of DNA abasic site protection by SRAP proteins
Abstract
Abasic (AP) sites are one of the most frequently occurring types of DNA damage. They lead to DNA strand breaks, interstrand DNA crosslinks, and block transcription and replication. Mutagenicity of AP sites arises from translesion synthesis (TLS) by error-prone bypass polymerases. Recently, a new cellular response to AP sites was discovered, in which the protein HMCES (5-hydroxymethlycytosine (5hmC) binding, embryonic stem cell-specific) forms a stable, covalent DNA-protein crosslink (DPC) to AP sites at stalled replication forks. The stability of the HMCES-DPC prevents strand cleavage by endonucleases and mutagenic bypass by TLS polymerases. Crosslinking is carried out by a unique SRAP (SOS Response Associated Peptidase) domain conserved across all domains of life. Here, we review the collection of recently reported SRAP crystal structures from human HMCES and E. coli YedK, which provide a unified basis for SRAP specificity and a putative chemical mechanism of AP site crosslinking. We discuss the structural and chemical basis for the stability of the SRAP DPC and how it differs from covalent protein-DNA intermediates in DNA lyase catalysis of strand scission.
Keywords: Abasic site; DNA lyase; DNA-protein crosslink; HMCES; SRAP; Thiazolidine.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
DECLARATION OF COMPETING INTEREST
There are no conflicts of interest to declare.
Figures

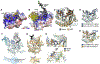
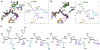

References
-
- Nakamura J, Walker VE, Upton PB, Chiang S-Y, Kow YW and Swenberg JA (1998) Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer research, 58, 222–225. - PubMed
-
- Nakamura J and Swenberg JA (1999) Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer research, 59, 2522–2526. - PubMed
-
- Lindahl T (1993) Instability and decay of the primary structure of DNA. nature, 362, 709–715. - PubMed
-
- Barnes DE and Lindahl T (2004) Repair and Genetic Consequences of Endogenous DNA Base Damage in Mammalian Cells. Annual Review of Genetics, 38, 445–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

