Nucleophosmin in Its Interaction with Ligands
- PMID: 32664415
- PMCID: PMC7402337
- DOI: 10.3390/ijms21144885
Nucleophosmin in Its Interaction with Ligands
Abstract
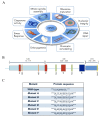
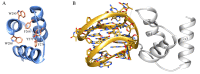
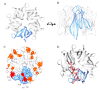
Nucleophosmin (NPM1) is a mainly nucleolar protein that shuttles between nucleoli, nucleoplasm and cytoplasm to fulfill its many functions. It is a chaperone of both nucleic acids and proteins and plays a role in cell cycle control, centrosome duplication, ribosome maturation and export, as well as the cellular response to a variety of stress stimuli. NPM1 is a hub protein in nucleoli where it contributes to nucleolar organization through heterotypic and homotypic interactions. Furthermore, several alterations, including overexpression, chromosomal translocations and mutations are present in solid and hematological cancers. Recently, novel germline mutations that cause dyskeratosis congenita have also been described. This review focuses on NPM1 interactions and inhibition. Indeed, the list of NPM1 binding partners is ever-growing and, in recent years, many studies contributed to clarifying the structural basis for NPM1 recognition of both nucleic acids and several proteins. Intriguingly, a number of natural and synthetic ligands that interfere with NPM1 interactions have also been reported. The possible role of NPM1 inhibitors in the treatment of multiple cancers and other pathologies is emerging as a new therapeutic strategy.
Keywords: AML; B23; acute myeloid leukemia; cancer; nucleophosmin; protein–nucleic acids interactions; protein–protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

