The staphylococcal biofilm protein Aap forms a tetrameric species as a necessary intermediate before amyloidogenesis
- PMID: 32665400
- PMCID: PMC7489908
- DOI: 10.1074/jbc.RA120.013936
The staphylococcal biofilm protein Aap forms a tetrameric species as a necessary intermediate before amyloidogenesis
Abstract
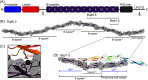
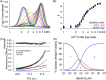
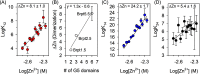
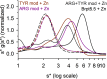
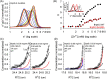
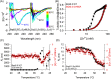
The accumulation-associated protein (Aap) from Staphylococcus epidermidis is a biofilm-related protein that was found to be a critical factor for infection using a rat catheter model. The B-repeat superdomain of Aap, composed of 5-17 B-repeats, each containing a Zn2+-binding G5 and a spacer subdomain, is responsible for Zn2+-dependent assembly leading to accumulation of bacteria during biofilm formation. We previously demonstrated that a minimal B-repeat construct (Brpt1.5) forms an antiparallel dimer in the presence of 2-3 Zn2+ ions. More recently, we have reported the presence of functional amyloid-like fibrils composed of Aap within S. epidermidis biofilms and demonstrated that a biologically relevant construct containing five and a half B-repeats (Brpt5.5) forms amyloid-like fibrils similar to those observed in the biofilm. In this study, we analyze the initial assembly events of the Brpt5.5 construct. Analytical ultracentrifugation was utilized to determine hydrodynamic parameters of reversibly associating species and to perform linked equilibrium studies. Linkage studies indicated a mechanism of Zn2+-induced dimerization similar to smaller constructs; however, Brpt5.5 dimers could then undergo further Zn2+-induced assembly into a previously uncharacterized tetramer. This led us to search for potential Zn2+-binding sites outside of the dimer interface. We developed a Brpt5.5 mutant that was unable to form the tetramer and was concordantly incapable of amyloidogenesis. CD and dynamic light scattering indicate that a conformational transition in the tetramer species is a critical step preceding amyloidogenesis. This mechanistic model for B-repeat assembly and amyloidogenesis provides new avenues for potential therapeutic targeting of staphylococcal biofilms.
Keywords: analytical ultracentrifugation; biofilm; biophysics; chemical modification; oligomerization; protein aggregation; protein chemical modification; sedimentation equilibrium; thermodynamics.
© 2020 Yarawsky and Herr.
Conflict of interest statement
Conflict of interest—A. B. H. serves as a Scientific Advisory Board member for Hoth Therapeutics, Inc., holds equity in Hoth Therapeutics and Chelexa BioSciences, LLC, and was a co-inventor on three patents broadly related to the subject matter of this work.
Figures


Similar articles
-
Assembly landscape of the complete B-repeat superdomain from Staphylococcus epidermidis strain 1457.Biophys J. 2025 Jan 21;124(2):363-378. doi: 10.1016/j.bpj.2024.12.011. Epub 2024 Dec 11. Biophys J. 2025. PMID: 39668565
-
The biofilm adhesion protein Aap from Staphylococcus epidermidis forms zinc-dependent amyloid fibers.J Biol Chem. 2020 Apr 3;295(14):4411-4427. doi: 10.1074/jbc.RA119.010874. Epub 2020 Feb 26. J Biol Chem. 2020. PMID: 32102851 Free PMC article.
-
Functional consequences of B-repeat sequence variation in the staphylococcal biofilm protein Aap: deciphering the assembly code.Biochem J. 2017 Feb 1;474(3):427-443. doi: 10.1042/BCJ20160675. Epub 2016 Nov 21. Biochem J. 2017. PMID: 27872164 Free PMC article.
-
Protein-based biofilm matrices in Staphylococci.Front Cell Infect Microbiol. 2014 Dec 10;4:171. doi: 10.3389/fcimb.2014.00171. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25540773 Free PMC article. Review.
-
Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects.Front Cell Infect Microbiol. 2015 Feb 10;5:7. doi: 10.3389/fcimb.2015.00007. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25713785 Free PMC article. Review.
Cited by
-
Strong non-ideality effects at low protein concentrations: considerations for elongated proteins.Eur Biophys J. 2023 Jul;52(4-5):427-438. doi: 10.1007/s00249-023-01648-x. Epub 2023 Apr 13. Eur Biophys J. 2023. PMID: 37055656 Free PMC article.
-
Solution Structural Studies of Pre-amyloid Oligomer States of the Biofilm Protein Aap.J Mol Biol. 2022 Aug 30;434(16):167708. doi: 10.1016/j.jmb.2022.167708. Epub 2022 Jun 28. J Mol Biol. 2022. PMID: 35777467 Free PMC article.
-
Assembly landscape of the complete B-repeat superdomain from Staphylococcus epidermidis strain 1457.Biophys J. 2025 Jan 21;124(2):363-378. doi: 10.1016/j.bpj.2024.12.011. Epub 2024 Dec 11. Biophys J. 2025. PMID: 39668565
-
Virulence Factors in Coagulase-Negative Staphylococci.Pathogens. 2021 Feb 4;10(2):170. doi: 10.3390/pathogens10020170. Pathogens. 2021. PMID: 33557202 Free PMC article. Review.
-
The staphylococcal biofilm protein Aap mediates cell-cell adhesion through mechanically distinct homophilic and lectin interactions.PNAS Nexus. 2022 Dec 2;1(5):pgac278. doi: 10.1093/pnasnexus/pgac278. eCollection 2022 Nov. PNAS Nexus. 2022. PMID: 36712378 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

