HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains
- PMID: 32665593
- PMCID: PMC7360574
- DOI: 10.1038/s41467-020-17256-8
HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains
Erratum in
-
Publisher Correction: HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains.Nat Commun. 2020 Nov 26;11(1):6165. doi: 10.1038/s41467-020-20152-w. Nat Commun. 2020. PMID: 33244008 Free PMC article.
Abstract
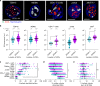
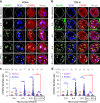
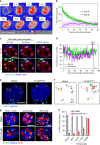
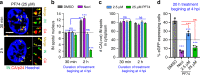
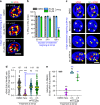
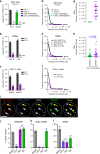
The early steps of HIV-1 infection, such as uncoating, reverse transcription, nuclear import, and transport to integration sites are incompletely understood. Here, we imaged nuclear entry and transport of HIV-1 replication complexes in cell lines, primary monocyte-derived macrophages (MDMs) and CD4+ T cells. We show that viral replication complexes traffic to and accumulate within nuclear speckles and that these steps precede the completion of viral DNA synthesis. HIV-1 transport to nuclear speckles is dependent on the interaction of the capsid proteins with host cleavage and polyadenylation specificity factor 6 (CPSF6), which is also required to stabilize the association of the viral replication complexes with nuclear speckles. Importantly, integration site analyses reveal a strong preference for HIV-1 to integrate into speckle-associated genomic domains. Collectively, our results demonstrate that nuclear speckles provide an architectural basis for nuclear homing of HIV-1 replication complexes and subsequent integration into associated genomic loci.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

