SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses
- PMID: 32665645
- PMCID: PMC7360742
- DOI: 10.1038/s41467-020-17488-8
SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses
Abstract
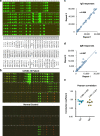
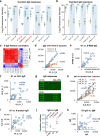
We still know very little about how the human immune system responds to SARS-CoV-2. Here we construct a SARS-CoV-2 proteome microarray containing 18 out of the 28 predicted proteins and apply it to the characterization of the IgG and IgM antibodies responses in the sera from 29 convalescent patients. We find that all these patients had IgG and IgM antibodies that specifically bind SARS-CoV-2 proteins, particularly the N protein and S1 protein. Besides these proteins, significant antibody responses to ORF9b and NSP5 are also identified. We show that the S1 specific IgG signal positively correlates with age and the level of lactate dehydrogenase (LDH) and negatively correlates with lymphocyte percentage. Overall, this study presents a systemic view of the SARS-CoV-2 specific IgG and IgM responses and provides insights to aid the development of effective diagnostic, therapeutic and vaccination strategies.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

