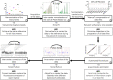
Standardization procedure for flow cytometry data harmonization in prospective multicenter studies
- PMID: 32665668
- PMCID: PMC7360585
- DOI: 10.1038/s41598-020-68468-3
Standardization procedure for flow cytometry data harmonization in prospective multicenter studies
Abstract
One of the most challenging objective for clinical cytometry in prospective multicenter immunomonitoring trials is to compare frequencies, absolute numbers of leukocyte populations and further the mean fluorescence intensities of cell markers, especially when the data are generated from different instruments. Here, we describe an innovative standardization workflow to compare all data to carry out any large-scale, prospective multicentric flow cytometry analysis whatever the duration, the number or type of instruments required for the realization of such projects.
Conflict of interest statement
M. H.-F. and J. M. are employees of UCB. A. B. and Z. M. are employees of BAYER Pharma. All other authors declare no competing financial and/or non-financial interests in relation to the work described.
Figures