A one-step, one-tube real-time RT-PCR based assay with an automated analysis for detection of SARS-CoV-2
- PMID: 32665985
- PMCID: PMC7341355
- DOI: 10.1016/j.heliyon.2020.e04405
A one-step, one-tube real-time RT-PCR based assay with an automated analysis for detection of SARS-CoV-2
Abstract
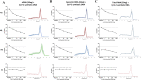
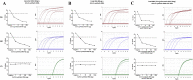
Early diagnosis of SARS-CoV-2 infected patients is essential to control the dynamics of the COVID-19 pandemic. We develop a rapid and accurate one-step multiplex TaqMan probe-based real-time RT-PCR assay, along with a computational tool to systematically analyse the data. Our assay could detect to a limit of 15 copies of SARS-CoV-2 transcripts-based on experiments performed by spiking total human RNA with in vitro synthesized viral transcripts. The assay was evaluated by performing 184 validations for the SARS-CoV-2 Nucleocapsid gene and human RNase P as an internal control reference gene with dilutions ranging from 1-100 ng for human RNA on a cohort of 26 clinical samples. 5 of 26 patients were confirmed to be infected with SARS-CoV-2, while 21 tested negative, consistent with the standards. The accuracy of the assay was found to be 100% sensitive and 100% specific based on the 26 clinical samples that need to be further verified using a large number of clinical samples. In summary, we present a rapid, easy to implement real-time PCR based assay with automated analysis using a novel COVID qPCR Analyzer tool with graphical user interface (GUI) to analyze the raw qRT-PCR data in an unbiased manner at a cost of under $3 per reaction and turnaround time of less than 2h, to enable in-house SARS-CoV-2 testing across laboratories.
Keywords: COVID qPCR analyzer; COVID-19 detection assay; Diagnostics; Genomics; Graphical user interface (GUI); Infectious disease; Microbial genomics; Molecular biology; Real-time RT- PCR; SARS-CoV-2; Virology.
© 2020 The Author(s).
Figures


Similar articles
-
Rapid, sensitive, and specific detection of SARS-CoV-2 in nasopharyngeal swab samples of suspected patients using a novel one-step loop-mediated isothermal amplification (one-step LAMP) technique.BMC Microbiol. 2023 Mar 7;23(1):63. doi: 10.1186/s12866-023-02806-z. BMC Microbiol. 2023. PMID: 36882699 Free PMC article.
-
Development and Validation of a Novel COVID-19 nsp8 One-Tube RT-LAMP-CRISPR Assay for SARS-CoV-2 Diagnosis.Microbiol Spectr. 2022 Dec 21;10(6):e0196222. doi: 10.1128/spectrum.01962-22. Epub 2022 Nov 29. Microbiol Spectr. 2022. PMID: 36445095 Free PMC article.
-
Automated SARS-COV-2 RNA extraction from patient nasopharyngeal samples using a modified DNA extraction kit for high throughput testing.Ann Saudi Med. 2020 Sep-Oct;40(5):373-381. doi: 10.5144/0256-4947.2020.373. Epub 2020 Oct 1. Ann Saudi Med. 2020. PMID: 32954791 Free PMC article.
-
Clinical evaluation of a multiplex real-time RT-PCR assay for detection of SARS-CoV-2 in individual and pooled upper respiratory tract samples.Arch Virol. 2021 Sep;166(9):2551-2561. doi: 10.1007/s00705-021-05148-1. Epub 2021 Jul 14. Arch Virol. 2021. PMID: 34259914 Free PMC article.
-
A Novel Multiplex qRT-PCR Assay to Detect SARS-CoV-2 Infection: High Sensitivity and Increased Testing Capacity.Microorganisms. 2020 Jul 17;8(7):1064. doi: 10.3390/microorganisms8071064. Microorganisms. 2020. PMID: 32708870 Free PMC article.
Cited by
-
Diagnostic techniques for COVID-19: A mini-review.J Virol Methods. 2022 Mar;301:114437. doi: 10.1016/j.jviromet.2021.114437. Epub 2021 Dec 18. J Virol Methods. 2022. PMID: 34933045 Free PMC article. Review.
-
Next-generation computational tools and resources for coronavirus research: From detection to vaccine discovery.Comput Biol Med. 2021 Jan;128:104158. doi: 10.1016/j.compbiomed.2020.104158. Epub 2020 Dec 1. Comput Biol Med. 2021. PMID: 33301953 Free PMC article. Review.
-
Emerging Multiplex Nucleic Acid Diagnostic Tests for Combating COVID-19.Biosensors (Basel). 2022 Nov 7;12(11):978. doi: 10.3390/bios12110978. Biosensors (Basel). 2022. PMID: 36354487 Free PMC article. Review.
-
Novel coronavirus disease (COVID-19) pandemic: A recent mini review.Comput Struct Biotechnol J. 2021;19:612-623. doi: 10.1016/j.csbj.2020.12.033. Epub 2020 Dec 31. Comput Struct Biotechnol J. 2021. PMID: 33398233 Free PMC article. Review.
-
Smart materials-integrated sensor technologies for COVID-19 diagnosis.Emergent Mater. 2021;4(1):169-185. doi: 10.1007/s42247-020-00150-w. Epub 2021 Jan 21. Emergent Mater. 2021. PMID: 33495747 Free PMC article. Review.
References
-
- Udugama B. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

