Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease
- PMID: 32668194
- PMCID: PMC7324317
- DOI: 10.1016/j.immuni.2020.06.024
Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease
Abstract
We profiled adaptive immunity in COVID-19 patients with active infection or after recovery and created a repository of currently >14 million B and T cell receptor (BCR and TCR) sequences from the blood of these patients. The B cell response showed converging IGHV3-driven BCR clusters closely associated with SARS-CoV-2 antibodies. Clonality and skewing of TCR repertoires were associated with interferon type I and III responses, early CD4+ and CD8+ T cell activation, and counterregulation by the co-receptors BTLA, Tim-3, PD-1, TIGIT, and CD73. Tfh, Th17-like, and nonconventional (but not classical antiviral) Th1 cell polarizations were induced. SARS-CoV-2-specific T cell responses were driven by TCR clusters shared between patients with a characteristic trajectory of clonotypes and traceability over the disease course. Our data provide fundamental insight into adaptive immunity to SARS-CoV-2 with the actively updated repository providing a resource for the scientific community urgently needed to inform therapeutic concepts and vaccine development.
Keywords: B cell repertoire; COVID-19; SARS-CoV-2-specific antibody; T cell compartments; T cell receptor clusters; T cell repertoire; cytokine profile; immunoglobulin heavy chain; interferon.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
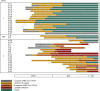
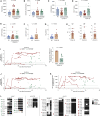




Comment on
-
COVID-19: consider cytokine storm syndromes and immunosuppression.Lancet. 2020 Mar 28;395(10229):1033-1034. doi: 10.1016/S0140-6736(20)30628-0. Epub 2020 Mar 16. Lancet. 2020. PMID: 32192578 Free PMC article. No abstract available.
References
-
- Becattini S., Latorre D., Mele F., Foglierini M., De Gregorio C., Cassotta A., Fernandez B., Kelderman S., Schumacher T.N., Corti D. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 2015;347:400–406. - PubMed
-
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

