High-Fat Diet and Antibiotics Cooperatively Impair Mitochondrial Bioenergetics to Trigger Dysbiosis that Exacerbates Pre-inflammatory Bowel Disease
- PMID: 32668218
- PMCID: PMC7429289
- DOI: 10.1016/j.chom.2020.06.001
High-Fat Diet and Antibiotics Cooperatively Impair Mitochondrial Bioenergetics to Trigger Dysbiosis that Exacerbates Pre-inflammatory Bowel Disease
Abstract
The clinical spectra of irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD) intersect to form a scantily defined overlap syndrome, termed pre-IBD. We show that increased Enterobacteriaceae and reduced Clostridia abundance distinguish the fecal microbiota of pre-IBD patients from IBS patients. A history of antibiotics in individuals consuming a high-fat diet was associated with the greatest risk for pre-IBD. Exposing mice to these risk factors resulted in conditions resembling pre-IBD and impaired mitochondrial bioenergetics in the colonic epithelium, which triggered dysbiosis. Restoring mitochondrial bioenergetics in the colonic epithelium with 5-amino salicylic acid, a PPAR-γ (peroxisome proliferator-activated receptor gamma) agonist that stimulates mitochondrial activity, ameliorated pre-IBD symptoms. As with patients, mice with pre-IBD exhibited notable expansions of Enterobacteriaceae that exacerbated low-grade mucosal inflammation, suggesting that remediating dysbiosis can alleviate inflammation. Thus, environmental risk factors cooperate to impair epithelial mitochondrial bioenergetics, thereby triggering microbiota disruptions that exacerbate inflammation and distinguish pre-IBD from IBS.
Keywords: antibiotics; dysbiosis; high-fat diet; inflammatory bowel disease; irritable bowel syndrome; microbiota.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

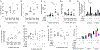
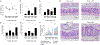

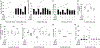

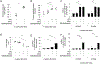
Comment in
-
Environmental stimuli and gut inflammation via dysbiosis in mouse and man.Nat Rev Gastroenterol Hepatol. 2020 Dec;17(12):715-716. doi: 10.1038/s41575-020-00373-6. Nat Rev Gastroenterol Hepatol. 2020. PMID: 33005024 No abstract available.
References
-
- Berrill JW, Green JT, Hood K, and Campbell AK (2013). Symptoms of irritable bowel syndrome in patients with inflammatory bowel disease: examining the role of sub-clinical inflammation and the impact on clinical assessment of disease activity. Aliment Pharmacol Ther 38, 44–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

