CeMbio - The Caenorhabditis elegans Microbiome Resource
- PMID: 32669368
- PMCID: PMC7466993
- DOI: 10.1534/g3.120.401309
CeMbio - The Caenorhabditis elegans Microbiome Resource
Abstract
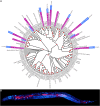
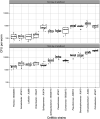
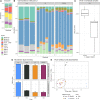
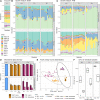
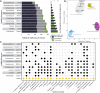
The study of microbiomes by sequencing has revealed a plethora of correlations between microbial community composition and various life-history characteristics of the corresponding host species. However, inferring causation from correlation is often hampered by the sheer compositional complexity of microbiomes, even in simple organisms. Synthetic communities offer an effective approach to infer cause-effect relationships in host-microbiome systems. Yet the available communities suffer from several drawbacks, such as artificial (thus non-natural) choice of microbes, microbe-host mismatch (e.g., human microbes in gnotobiotic mice), or hosts lacking genetic tractability. Here we introduce CeMbio, a simplified natural Caenorhabditis elegans microbiota derived from our previous meta-analysis of the natural microbiome of this nematode. The CeMbio resource is amenable to all strengths of the C. elegans model system, strains included are readily culturable, they all colonize the worm gut individually, and comprise a robust community that distinctly affects nematode life-history. Several tools have additionally been developed for the CeMbio strains, including diagnostic PCR primers, completely sequenced genomes, and metabolic network models. With CeMbio, we provide a versatile resource and toolbox for the in-depth dissection of naturally relevant host-microbiome interactions in C. elegans.
Keywords: C. elegans; Host-microbe interactions; Metabolic networks; Microbiome resource; Synthetic communities.
Copyright © 2020 Dirksen et al.
Figures

References
-
- Barrière A, M. A. Félix Isolation of C. elegans and related nematodes. WormBook. 2006: 1–9. https://doi:10.1895/wormbook.1.115.1 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
