Human Cardiac Mesenchymal Stromal Cells From Right and Left Ventricles Display Differences in Number, Function, and Transcriptomic Profile
- PMID: 32670081
- PMCID: PMC7327120
- DOI: 10.3389/fphys.2020.00604
Human Cardiac Mesenchymal Stromal Cells From Right and Left Ventricles Display Differences in Number, Function, and Transcriptomic Profile
Abstract
Background: Left ventricle (LV) and right ventricle (RV) are characterized by well-known physiological differences, mainly related to their different embryological origin, hemodynamic environment, function, structure, and cellular composition. Nevertheless, scarce information is available about cellular peculiarities between left and right ventricular chambers in physiological and pathological contexts. Cardiac mesenchymal stromal cells (C-MSC) are key cells affecting many functions of the heart. Differential features that distinguish LV from RV C-MSC are still underappreciated.
Aim: To analyze the physiological differential amount, function, and transcriptome of human C-MSC in LV versus (vs.) RV.
Methods: Human cardiac specimens of LV and RV from healthy donors were used for tissue analysis of C-MSC number, and for C-MSC isolation. Paired LV and RV C-MSC were compared as for surface marker expression, cell proliferation/death ratio, migration, differentiation capabilities, and transcriptome profile.
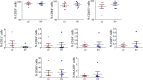
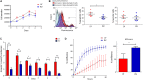
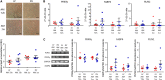
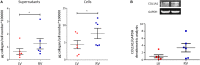
Results: Histological analysis showed a greater percentage of C-MSC in RV vs. LV tissue. Moreover, a higher C-MSC amount was obtained from RV than from LV after isolation procedures. LV and RV C-MSC are characterized by a similar proportion of surface markers. Functional studies revealed comparable cell growth curves in cells from both ventricles. Conversely, LV C-MSC displayed a higher apoptosis rate and RV C-MSC were characterized by a higher migration speed and collagen deposition. Consistently, transcriptome analysis showed that genes related to apoptosis regulation or extracellular matrix organization and integrins were over-expressed in LV and RV, respectively. Besides, we revealed additional pathways specifically associated with LV or RV C-MSC, including energy metabolism, inflammatory response, cardiac conduction, and pluripotency.
Conclusion: Taken together, these results contribute to the functional characterization of RV and LV C-MSC in physiological conditions. This information suggests a possible differential role of the stromal compartment in chamber-specific pathologic scenarios.
Keywords: cardiac mesenchymal stromal cells; cardiac ventricles; functional studies; left ventricle; right ventricle; transcriptome.
Copyright © 2020 Stadiotti, Piacentini, Vavassori, Chiesa, Scopece, Guarino, Micheli, Polvani, Colombo, Pompilio and Sommariva.
Figures



References
-
- Andersen S., Nielsen-Kudsk J. E., Vonk Noordegraaf A., de Man F. S. (2019). Right ventricular fibrosis. Circulation 139 269–285. - PubMed
-
- Banerjee I., Fuseler J. W., Price R. L., Borg T. K., Baudino T. A. (2007). Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiol. Heart Circ. Physiol. 293 H1883–H1891. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

