Engineering Escherichia coli to improve tryptophan production via genetic manipulation of precursor and cofactor pathways
- PMID: 32671235
- PMCID: PMC7334480
- DOI: 10.1016/j.synbio.2020.06.009
Engineering Escherichia coli to improve tryptophan production via genetic manipulation of precursor and cofactor pathways
Abstract
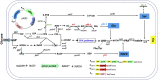
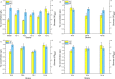
Optimizing the supply of biosynthetic precursors and cofactors is usually an effective metabolic strategy to improve the production of target compounds. Here, the combination of optimizing precursor synthesis and balancing cofactor metabolism was adopted to improve tryptophan production in Escherichia coli. First, glutamine synthesis was improved by expressing heterologous glutamine synthetase from Bacillus subtilis and Bacillus megaterium in the engineered Escherichia coli strain KW001, resulting in the best candidate strain TS-1. Then icd and gdhA were overexpressed in TS-1, which led to the accumulation of 1.060 g/L tryptophan. Subsequently, one more copy of prs was introduced on the chromosome to increase the flux of 5-phospho-α-d-ribose 1-diphosphate followed by the expression of mutated serA and thrA to increase the precursor supply of serine, resulting in the accumulation of 1.380 g/L tryptophan. Finally, to maintain cofactor balance, sthA and pntAB, encoding transhydrogenase, were overexpressed. With sufficient amounts of precursors and balanced cofactors, the engineered strain could produce 1.710 g/L tryptophan after 48 h of shake-flask fermentation, which was 2.76-times higher than the titer of the parent strain. Taken together, our results demonstrate that the combination of optimizing precursor supply and regulating cofactor metabolism is an effective approach for high-level production of tryptophan. Similar strategies could be applied to the production of other amino acids or related derivatives.
Keywords: Cofactor supply; Escherichia coli; Metabolic precursors; Tryptophan.
© 2020 Production and hosting by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Figures
References
-
- Won-Gi Bang S.L., Hermann Sahm T., Fritz Wagner. Production of L-tryptophan by Escherichia coli cells. Cell. 1983;XXV:999–1011. - PubMed
-
- Zhao Z.J., Zou C., Zhu Y.X., Dai J., Chen S., Wu D. Development of L-tryptophan production strains by defined genetic modification in Escherichia coli. J Ind Microbiol Biotechnol. 2011;38:1921–1929. - PubMed
-
- Ikeda M., Katsumata R. Tryptophan production by transport mutants of Corynebacterium glutamicum. Biosci Biotechnol Biochem. 1995;59:1600–1602.
LinkOut - more resources
Full Text Sources
Other Literature Sources

