Insights into epigenetic patterns in mammalian early embryos
- PMID: 32671792
- PMCID: PMC7815849
- DOI: 10.1007/s13238-020-00757-z
Insights into epigenetic patterns in mammalian early embryos
Abstract
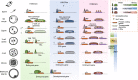
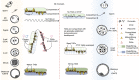
Mammalian fertilization begins with the fusion of two specialized gametes, followed by major epigenetic remodeling leading to the formation of a totipotent embryo. During the development of the pre-implantation embryo, precise reprogramming progress is a prerequisite for avoiding developmental defects or embryonic lethality, but the underlying molecular mechanisms remain elusive. For the past few years, unprecedented breakthroughs have been made in mapping the regulatory network of dynamic epigenomes during mammalian early embryo development, taking advantage of multiple advances and innovations in low-input genome-wide chromatin analysis technologies. The aim of this review is to highlight the most recent progress in understanding the mechanisms of epigenetic remodeling during early embryogenesis in mammals, including DNA methylation, histone modifications, chromatin accessibility and 3D chromatin organization.
Keywords: DNA methylation; early embryo development; epigenetic reprogramming; histone modifications.
Conflict of interest statement
Ruimin Xu, Chong Li, Xiaoyu Liu and Shaorong Gao declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
Figures

Similar articles
-
Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment.Trends Cell Biol. 2018 Mar;28(3):237-253. doi: 10.1016/j.tcb.2017.10.008. Epub 2017 Dec 5. Trends Cell Biol. 2018. PMID: 29217127 Review.
-
Genomic insights into chromatin reprogramming to totipotency in embryos.J Cell Biol. 2019 Jan 7;218(1):70-82. doi: 10.1083/jcb.201807044. Epub 2018 Sep 26. J Cell Biol. 2019. PMID: 30257850 Free PMC article. Review.
-
Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals.Mol Cell. 2016 Sep 15;63(6):1066-79. doi: 10.1016/j.molcel.2016.08.032. Mol Cell. 2016. PMID: 27635762
-
Epigenetic Regulation and Risk Factors During the Development of Human Gametes and Early Embryos.Annu Rev Genomics Hum Genet. 2019 Aug 31;20:21-40. doi: 10.1146/annurev-genom-083118-015143. Epub 2019 Mar 27. Annu Rev Genomics Hum Genet. 2019. PMID: 30917080 Review.
-
Genome-wide profiling of the epigenetic landscape of histone variant TH2B in murine oocytes and pre-implantation embryos.Reproduction. 2025 Jan 2;169(1):e240035. doi: 10.1530/REP-24-0035. Print 2025 Jan 1. Reproduction. 2025. PMID: 39447008
Cited by
-
Epigenetics Mechanisms of Honeybees: Secrets of Royal Jelly.Epigenet Insights. 2023 Nov 29;16:25168657231213717. doi: 10.1177/25168657231213717. eCollection 2023. Epigenet Insights. 2023. PMID: 38033464 Free PMC article. Review.
-
The Genetic and Epigenetic Toxicity of Silica Nanoparticles: An Updated Review.Int J Nanomedicine. 2024 Dec 24;19:13901-13923. doi: 10.2147/IJN.S486858. eCollection 2024. Int J Nanomedicine. 2024. PMID: 39735322 Free PMC article. Review.
-
TDG is a pig-specific epigenetic regulator with insensitivity to H3K9 and H3K27 demethylation in nuclear transfer embryos.Stem Cell Reports. 2021 Nov 9;16(11):2674-2689. doi: 10.1016/j.stemcr.2021.09.012. Epub 2021 Oct 21. Stem Cell Reports. 2021. PMID: 34678203 Free PMC article.
-
Aberrant KAT2A accumulations render TRIM22-low melanoma sensitive to Notch1 inhibitors via epigenetic reprogramming.J Transl Med. 2023 Jul 6;21(1):443. doi: 10.1186/s12967-023-04305-1. J Transl Med. 2023. PMID: 37415153 Free PMC article.
-
Epigenetic implications in maternal diabetes and metabolic syndrome-associated risk of orofacial clefts.Birth Defects Res. 2023 Nov 15;115(19):1835-1850. doi: 10.1002/bdr2.2226. Epub 2023 Jul 27. Birth Defects Res. 2023. PMID: 37497595 Free PMC article. Review.
References
-
- Amdani SN, Yeste M, Jones C, Coward K. Sperm factors and oocyte activation: current controversies and considerations. Biol Reprod. 2015;93:50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

