JAM-A Acts via C/EBP-α to Promote Claudin-5 Expression and Enhance Endothelial Barrier Function
- PMID: 32673519
- PMCID: PMC7508279
- DOI: 10.1161/CIRCRESAHA.120.316742
JAM-A Acts via C/EBP-α to Promote Claudin-5 Expression and Enhance Endothelial Barrier Function
Abstract
Rationale: Intercellular tight junctions are crucial for correct regulation of the endothelial barrier. Their composition and integrity are affected in pathological contexts, such as inflammation and tumor growth. JAM-A (junctional adhesion molecule A) is a transmembrane component of tight junctions with a role in maintenance of endothelial barrier function, although how this is accomplished remains elusive.
Objective: We aimed to understand the molecular mechanisms through which JAM-A expression regulates tight junction organization to control endothelial permeability, with potential implications under pathological conditions.
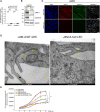
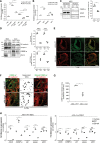
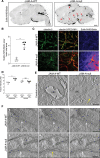
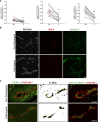
Methods and results: Genetic deletion of JAM-A in mice significantly increased vascular permeability. This was associated with significantly decreased expression of claudin-5 in the vasculature of various tissues, including brain and lung. We observed that C/EBP-α (CCAAT/enhancer-binding protein-α) can act as a transcription factor to trigger the expression of claudin-5 downstream of JAM-A, to thus enhance vascular barrier function. Accordingly, gain-of-function for C/EBP-α increased claudin-5 expression and decreased endothelial permeability, as measured by the passage of fluorescein isothiocyanate (FITC)-dextran through endothelial monolayers. Conversely, C/EBP-α loss-of-function showed the opposite effects of decreased claudin-5 levels and increased endothelial permeability. Mechanistically, JAM-A promoted C/EBP-α expression through suppression of β-catenin transcriptional activity, and also through activation of EPAC (exchange protein directly activated by cAMP). C/EBP-α then directly binds the promoter of claudin-5 to thereby promote its transcription. Finally, JAM-A-C/EBP-α-mediated regulation of claudin-5 was lost in blood vessels from tissue biopsies from patients with glioblastoma and ovarian cancer.
Conclusions: We describe here a novel role for the transcription factor C/EBP-α that is positively modulated by JAM-A, a component of tight junctions that acts through EPAC to up-regulate the expression of claudin-5, to thus decrease endothelial permeability. Overall, these data unravel a regulatory molecular pathway through which tight junctions limit vascular permeability. This will help in the identification of further therapeutic targets for diseases associated with endothelial barrier dysfunction. Graphic Abstract: An graphic abstract is available for this article.
Keywords: claudin-5; endothelium; junctional adhesion molecule A; tight junctions; vascular permeability.
Conflict of interest statement
None.
Figures


References
-
- Mehta D, Malik AB. Signaling mechanisms regulating endothelial permeability. Physiol Rev. 2006;86:279–367. doi: 10.1152/physrev.00012.2005 - PubMed
-
- Dejana E. Endothelial cell-cell junctions: happy together. Nat Rev Mol Cell Biol. 2004;5:261–270. doi: 10.1038/nrm1357 - PubMed
-
- Bazzoni G, Dejana E. Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol Rev. 2004;84:869–901. doi: 10.1152/physrev.00035.2003 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

