Activating Transcription Factor 6 Mediates Inflammatory Signals in Intestinal Epithelial Cells Upon Endoplasmic Reticulum Stress
- PMID: 32673694
- PMCID: PMC7923714
- DOI: 10.1053/j.gastro.2020.06.088
Activating Transcription Factor 6 Mediates Inflammatory Signals in Intestinal Epithelial Cells Upon Endoplasmic Reticulum Stress
Abstract
Background & aims: Excess and unresolved endoplasmic reticulum (ER) stress in intestinal epithelial cells (IECs) promotes intestinal inflammation. Activating transcription factor 6 (ATF6) is one of the signaling mediators of ER stress. We studied the pathways that regulate ATF6 and its role for inflammation in IECs.
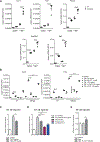
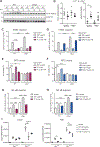
Methods: We performed an RNA interference screen, using 23,349 unique small interfering RNAs targeting 7783 genes and a luciferase reporter controlled by an ATF6-dependent ERSE (ER stress-response element) promoter, to identify proteins that activate or inhibit the ATF6 signaling pathway in HEK293 cells. To validate the screening results, intestinal epithelial cell lines (Caco-2 cells) were transfected with small interfering RNAs or with a plasmid overexpressing a constitutively active form of ATF6. Caco-2 cells with a CRISPR-mediated disruption of autophagy related 16 like 1 gene (ATG16L1) were used to study the effect of ATF6 on ER stress in autophagy-deficient cells. We also studied intestinal organoids derived from mice that overexpress constitutively active ATF6, from mice with deletion of the autophagy related 16 like 1 or X-Box binding protein 1 gene in IECs (Atg16l1ΔIEC or Xbp1ΔIEC, which both develop spontaneous ileitis), from patients with Crohn's disease (CD) and healthy individuals (controls). Cells and organoids were incubated with tunicamycin to induce ER stress and/or chemical inhibitors of newly identified activator proteins of ATF6 signaling, and analyzed by real-time polymerase chain reaction and immunoblots. Atg16l1ΔIEC and control (Atg16l1fl/fl) mice were given intraperitoneal injections of tunicamycin and were treated with chemical inhibitors of ATF6 activating proteins.
Results: We identified and validated 15 suppressors and 7 activators of the ATF6 signaling pathway; activators included the regulatory subunit of casein kinase 2 (CSNK2B) and acyl-CoA synthetase long chain family member 1 (ACSL1). Knockdown or chemical inhibition of CSNK2B and ACSL1 in Caco-2 cells reduced activity of the ATF6-dependent ERSE reporter gene, diminished transcription of the ATF6 target genes HSP90B1 and HSPA5 and reduced NF-κB reporter gene activation on tunicamycin stimulation. Atg16l1ΔIEC and or Xbp1ΔIEC organoids showed increased expression of ATF6 and its target genes. Inhibitors of ACSL1 or CSNK2B prevented activation of ATF6 and reduced CXCL1 and tumor necrosis factor (TNF) expression in these organoids on induction of ER stress with tunicamycin. Injection of mice with inhibitors of ACSL1 or CSNK2B significantly reduced tunicamycin-mediated intestinal inflammation and IEC death and expression of CXCL1 and TNF in Atg16l1ΔIEC mice. Purified ileal IECs from patients with CD had higher levels of ATF6, CSNK2B, and HSPA5 messenger RNAs than controls; early-passage organoids from patients with active CD show increased levels of activated ATF6 protein, incubation of these organoids with inhibitors of ACSL1 or CSNK2B reduced transcription of ATF6 target genes, including TNF.
Conclusions: Ileal IECs from patients with CD have higher levels of activated ATF6, which is regulated by CSNK2B and HSPA5. ATF6 increases expression of TNF and other inflammatory cytokines in response to ER stress in these cells and in organoids from Atg16l1ΔIEC and Xbp1ΔIEC mice. Strategies to inhibit the ATF6 signaling pathway might be developed for treatment of inflammatory bowel diseases.
Keywords: Gene Expression; IBD; Inflammation; Signal Transduction.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





References
-
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999;397:271–4. - PubMed
-
- Yoshida H, Haze K, Yanagi H, et al. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem 1998;273:33741–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

