Genetic variation between long-lived versus short-lived bats illuminates the molecular signatures of longevity
- PMID: 32674072
- PMCID: PMC7485743
- DOI: 10.18632/aging.103725
Genetic variation between long-lived versus short-lived bats illuminates the molecular signatures of longevity
Abstract
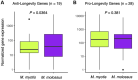
Bats are the longest-lived mammals given their body size with majority of species exhibiting exceptional longevity. However, there are some short-lived species that do not exhibit extended lifespans. Here we conducted a comparative genomic and transcriptomic study on long-lived Myotis myotis (maximum lifespan = 37.1 years) and short-lived Molossus molossus (maximum lifespan = 5.6 years) to ascertain the genetic difference underlying their divergent longevities. Genome-wide selection tests on 12,467 single-copy genes between M. myotis and M. molossus revealed only three genes (CCDC175, FATE1 and MLKL) that exhibited significant positive selection. Although 97.96% of 12,467 genes underwent purifying selection, we observed a significant heterogeneity in their expression patterns. Using a linear mixed model, we obtained expression of 2,086 genes that may truly represent the genetic difference between M. myotis and M. molossus. Expression analysis indicated that long-lived M. myotis exhibited a transcriptomic profile of enhanced DNA repair and autophagy pathways, compared to M. molossus. Further investigation of the longevity-associated genes suggested that long-lived M. myotis have naturally evolved a diminished anti-longevity transcriptomic profile. Together with observations from other long-lived species, our results suggest that heightened DNA repair and autophagy activity may represent a universal mechanism to achieve longevity in long-lived mammals.
Keywords: autophagy; bats; comparative genomics; longevity; transcriptomics.
Conflict of interest statement
Figures




References
-
- George JC, Bada J, Zeh J, Scott L, Brown SE, O'Hara T, Suydam R. Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can J Zool. 1999; 77:571–580. 10.1139/z99-015 - DOI
-
- Foley NM, Hughes GM, Huang Z, Clarke M, Jebb D, Whelan CV, Petit EJ, Touzalin F, Farcy O, Jones G, Ransome RD, Kacprzyk J, O’Connell MJ, et al. Growing old, yet staying young: the role of telomeres in bats’ exceptional longevity. Sci Adv. 2018; 4:eaao0926. 10.1126/sciadv.aao0926 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

