Modeling Approaches Reveal New Regulatory Networks in Aspergillus fumigatus Metabolism
- PMID: 32674323
- PMCID: PMC7557846
- DOI: 10.3390/jof6030108
Modeling Approaches Reveal New Regulatory Networks in Aspergillus fumigatus Metabolism
Abstract
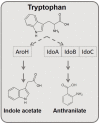
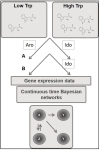
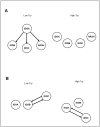
Systems biology approaches are extensively used to model and reverse-engineer gene regulatory networks from experimental data. Indoleamine 2,3-dioxygenases (IDOs)-belonging in the heme dioxygenase family-degrade l-tryptophan to kynurenines. These enzymes are also responsible for the de novo synthesis of nicotinamide adenine dinucleotide (NAD+). As such, they are expressed by a variety of species, including fungi. Interestingly, Aspergillus may degrade l-tryptophan not only via IDO but also via alternative pathways. Deciphering the molecular interactions regulating tryptophan metabolism is particularly critical for novel drug target discovery designed to control pathogen determinants in invasive infections. Using continuous time Bayesian networks over a time-course gene expression dataset, we inferred the global regulatory network controlling l-tryptophan metabolism. The method unravels a possible novel approach to target fungal virulence factors during infection. Furthermore, this study represents the first application of continuous-time Bayesian networks as a gene network reconstruction method in Aspergillus metabolism. The experiment showed that the applied computational approach may improve the understanding of metabolic networks over traditional pathways.
Keywords: Aspergillus fumigatus; Bayesian networks; continuous time Bayesian networks; gene network inference; gene network reconstruction; modeling; tryptophan metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Grants and funding
- PGR13XNIDJ/The Italian Grant "Programma per Giovani Ricercatori - Rita Levi Montalcini 2013
- Project MAGNET CZ.02.1.01/0.0/0.0/15_003/0000492/European Social Fund and European Regional Development Fund
- ENOCH CZ.02.1.01/0.0/0.0/16_019/0000868/European Social Fund and European Regional Development Fund
- NV18-06-00529/Ministry of Health of the Czech Republic
LinkOut - more resources
Full Text Sources
Research Materials

