Phylogenetic Analyses of Rotavirus A from Cattle in Uruguay Reveal the Circulation of Common and Uncommon Genotypes and Suggest Interspecies Transmission
- PMID: 32674420
- PMCID: PMC7400708
- DOI: 10.3390/pathogens9070570
Phylogenetic Analyses of Rotavirus A from Cattle in Uruguay Reveal the Circulation of Common and Uncommon Genotypes and Suggest Interspecies Transmission
Abstract
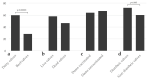
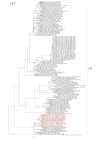
Uruguay is one of the main exporters of beef and dairy products, and cattle production is one of the main economic sectors in this country. Rotavirus A (RVA) is the main pathogen associated with neonatal calf diarrhea (NCD), a syndrome that leads to significant economic losses to the livestock industry. The aims of this study are to determine the frequency of RVA infections, and to analyze the genetic diversity of RVA strains in calves in Uruguay. A total of 833 samples from dairy and beef calves were analyzed through RT-qPCR and sequencing. RVA was detected in 57.0% of the samples. The frequency of detection was significantly higher in dairy (59.5%) than beef (28.4%) calves (p < 0.001), while it did not differ significantly among calves born in herds that were vaccinated (64.0%) or not vaccinated (66.7%) against NCD. The frequency of RVA detection and the viral load were significantly higher in samples from diarrheic (72.1%, 7.99 log10 genome copies/mL of feces) than non-diarrheic (59.9%, 7.35 log10 genome copies/mL of feces) calves (p < 0.005 and p = 0.007, respectively). The observed G-types (VP7) were G6 (77.6%), G10 (20.7%), and G24 (1.7%), while the P-types were P[5] (28.4%), P[11] (70.7%), and P[33] (0.9%). The G-type and P-type combinations were G6P[11] (40.4%), G6P[5] (38.6%), G10P[11] (19.3%), and the uncommon genotype G24P[33] (1.8%). VP6 and NSP1-5 genotyping were performed to better characterize some strains. The phylogenetic analyses suggested interspecies transmission, including transmission between animals and humans.
Keywords: bovine; diarrhea; genotypes; interspecies transmission; rotavirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Urie N.J., Lombard J.E., Shivley C.B., Kopral C.A., Adams A.E., Earleywine T.J., Olson J.D., Garry F.B. Preweaned heifer management on US dairy operations: Part V. Factors associated with morbidity and mortality in preweaned dairy heifer calves. J. Dairy Sci. 2018;101:9229–9244. doi: 10.3168/jds.2017-14019. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

