Stem Cell Pluripotency Genes Klf4 and Oct4 Regulate Complex SMC Phenotypic Changes Critical in Late-Stage Atherosclerotic Lesion Pathogenesis
- PMID: 32674599
- PMCID: PMC7682794
- DOI: 10.1161/CIRCULATIONAHA.120.046672
Stem Cell Pluripotency Genes Klf4 and Oct4 Regulate Complex SMC Phenotypic Changes Critical in Late-Stage Atherosclerotic Lesion Pathogenesis
Abstract
Background: Rupture and erosion of advanced atherosclerotic lesions with a resultant myocardial infarction or stroke are the leading worldwide cause of death. However, we have a limited understanding of the identity, origin, and function of many cells that make up late-stage atherosclerotic lesions, as well as the mechanisms by which they control plaque stability.
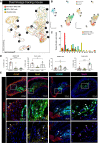
Methods: We conducted a comprehensive single-cell RNA sequencing of advanced human carotid endarterectomy samples and compared these with single-cell RNA sequencing from murine microdissected advanced atherosclerotic lesions with smooth muscle cell (SMC) and endothelial lineage tracing to survey all plaque cell types and rigorously determine their origin. We further used chromatin immunoprecipitation sequencing (ChIP-seq), bulk RNA sequencing, and an innovative dual lineage tracing mouse to understand the mechanism by which SMC phenotypic transitions affect lesion pathogenesis.
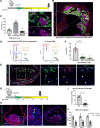
Results: We provide evidence that SMC-specific Klf4- versus Oct4-knockout showed virtually opposite genomic signatures, and their putative target genes play an important role regulating SMC phenotypic changes. Single-cell RNA sequencing revealed remarkable similarity of transcriptomic clusters between mouse and human lesions and extensive plasticity of SMC- and endothelial cell-derived cells including 7 distinct clusters, most negative for traditional markers. In particular, SMC contributed to a Myh11-, Lgals3+ population with a chondrocyte-like gene signature that was markedly reduced with SMC-Klf4 knockout. We observed that SMCs that activate Lgals3 compose up to two thirds of all SMC in lesions. However, initial activation of Lgals3 in these cells does not represent conversion to a terminally differentiated state, but rather represents transition of these cells to a unique stem cell marker gene-positive, extracellular matrix-remodeling, "pioneer" cell phenotype that is the first to invest within lesions and subsequently gives rise to at least 3 other SMC phenotypes within advanced lesions, including Klf4-dependent osteogenic phenotypes likely to contribute to plaque calcification and plaque destabilization.
Conclusions: Taken together, these results provide evidence that SMC-derived cells within advanced mouse and human atherosclerotic lesions exhibit far greater phenotypic plasticity than generally believed, with Klf4 regulating transition to multiple phenotypes including Lgals3+ osteogenic cells likely to be detrimental for late-stage atherosclerosis plaque pathogenesis.
Keywords: coronary disease; single nucleotide polymorphisms; smooth muscle cell; smooth muscle differentiation; smooth muscle progenitor cell.
Conflict of interest statement
None.
Figures




References
-
- Bentzon JF, Otsuka F, Virmani R, Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014; 114:1852–1866. doi: 10.1161/CIRCRESAHA.114.302721 - PubMed
-
- Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011; 473:317–325. doi: 10.1038/nature10146 - PubMed
-
- Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015; 21:628–637. doi: 10.1038/nm.3866 - PMC - PubMed
-
- Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res. 2014; 115:662–667. doi: 10.1161/CIRCRESAHA.115.304634 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

