The contribution of hereditary cancer-related germline mutations to lung cancer susceptibility
- PMID: 32676327
- PMCID: PMC7354149
- DOI: 10.21037/tlcr-19-403
The contribution of hereditary cancer-related germline mutations to lung cancer susceptibility
Abstract
Background: Germline variations may contribute to lung cancer susceptibility besides environmental factors. The influence of germline mutations on lung cancer susceptibility and their correlation with somatic mutations has not been systematically investigated.
Methods: In this study, germline mutations from 1,026 non-small cell lung cancer (NSCLC) patients were analyzed with a 58-gene next-generation sequencing (NGS) panel containing known hereditary cancer-related genes, and were categorized based on American College of Medical Genetics and Genomics (ACMG) guidelines in pathogenicity, and the corresponding somatic mutations were analyzed using a 605-gene NGS panel containing known cancer-related genes.
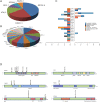
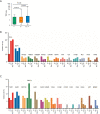
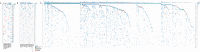
Results: Plausible genetic susceptibility was found in 4.7% of lung cancer patients, in which 14 patients with pathogenic mutations (P group) and 34 patients with likely-pathogenic mutations (LP group) were identified. The ratio of the first degree relatives with lung cancer history of the P groups was significantly higher than the Non-P group (P=0.009). The ratio of lung cancer patients with history of other cancers was higher in P (P=0.0007) or LP (P=0.017) group than the Non-P group. Pathogenic mutations fell most commonly in BRCA2, followed by CHEK2 and ATM. Likely-pathogenic mutations fell most commonly in NTRK1 and EXT2, followed by BRIP1 and PALB2. These genes are involved in DNA repair, cell cycle regulation and tumor suppression. By comparing the germline mutation frequency from this study with that from the whole population or East Asian population (gnomAD database), we found that the overall odds ratio (OR) for P or LP group was 17.93 and 15.86, respectively, when compared with the whole population, and was 2.88 and 3.80, respectively, when compared with the East Asian population, suggesting the germline mutations of the P and LP groups were risk factors for lung cancer. Somatic mutation analysis revealed no significant difference in tumor mutation burden (TMB) among the groups, although a trend of lower TMB in the pathogenic group was found. The SNV/INDEL mutation frequency of TP53 in the P group was significantly lower than the other two groups, and the copy number variation (CNV) mutation frequency of PIK3CA and MET was significantly higher than the Non-P group. Pathway enrichment analysis found no significant difference in aberrant pathways among the three groups.
Conclusions: A proportion of 4.7% of patients carrying germline variants may be potentially linked to increased susceptibility to lung cancer. Patients with pathogenic germline mutations exhibited stronger family history and higher lung cancer risk.
Keywords: BRCA2; EGFR; Lung cancer; germline; pathogenic; susceptibility.
2020 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-403). Mengyuan L, XL, PS, TH, YZ, Ming L, LL, Yaru C, YZ, GL, JY and SC report non-financial support from HaploX Biotechnology outside the submitted work. LS reports grants from The Special Funds for Strategic Emerging Industry Development of Shenzhen and The Science and Technology Project of Shenzhen, non-financial support and other from HaploX Biotechnology Co., Ltd. outside the submitted work. The other authors have no conflicts of interest to declare.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
