A large-scale genomic association analysis identifies the candidate causal genes conferring stripe rust resistance under multiple field environments
- PMID: 32677132
- PMCID: PMC7769225
- DOI: 10.1111/pbi.13452
A large-scale genomic association analysis identifies the candidate causal genes conferring stripe rust resistance under multiple field environments
Abstract
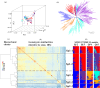
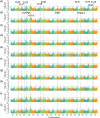
The incorporation of resistance genes into wheat commercial varieties is the ideal strategy to combat stripe or yellow rust (YR). In a search for novel resistance genes, we performed a large-scale genomic association analysis with high-density 660K single nucleotide polymorphism (SNP) arrays to determine the genetic components of YR resistance in 411 spring wheat lines. Following quality control, 371 972 SNPs were screened, covering over 50% of the high-confidence annotated gene space. Nineteen stable genomic regions harbouring 292 significant SNPs were associated with adult-plant YR resistance across nine environments. Of these, 14 SNPs were localized in the proximity of known loci widely used in breeding. Obvious candidate SNP variants were identified in certain confidence intervals, such as the cloned gene Yr18 and the major locus on chromosome 2BL, despite a large extent of linkage disequilibrium. The number of causal SNP variants was refined using an independent validation panel and consideration of the estimated functional importance of each nucleotide polymorphism. Interestingly, four natural polymorphisms causing amino acid changes in the gene TraesCS2B01G513100 that encodes a serine/threonine protein kinase (STPK) were significantly involved in YR responses. Gene expression and mutation analysis confirmed that STPK played an important role in YR resistance. PCR markers were developed to identify the favourable TraesCS2B01G513100 haplotype for marker-assisted breeding. These results demonstrate that high-resolution SNP-based GWAS enables the rapid identification of putative resistance genes and can be used to improve the efficiency of marker-assisted selection in wheat disease resistance breeding.
Keywords: 660K SNP array; GWAS; candidate region association analysis; common wheat (Triticum aestivum L.); marker-assisted breeding; serine/threonine protein kinase (STPK); stripe rust.
© 2020 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest and all experiments comply with the current laws of China.
Figures



References
-
- Arora, S. , Steuernagel, B. , Gaurav, K. , Chandramohan, S. , Long, Y. , Matny, O. , Johnson, R. et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143. - PubMed
-
- Bates, D. , Maechler, M. , Bolker, B. and Walker, S. (2014) lme4: linear mixed‐effects models using Eigen and S4. pp 1–23.
-
- Bazakos, C. , Hanemian, M. , Trontin, C. , Jimenez‐Gomez, J.M. and Loudet, O. (2017) New strategies and tools in quantitative genetics: how to go from the phenotype to the genotype. Annu. Rev. Plant Biol. 68, 435–455. - PubMed
-
- Brown, J.K. (2015) Durable resistance of crops to disease: a Darwinian perspective. Annu. Rev. Phytopathol. 53, 513–539. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

