Epigenomic programming in early fetal brain development
- PMID: 32677466
- PMCID: PMC7857341
- DOI: 10.2217/epi-2019-0319
Epigenomic programming in early fetal brain development
Abstract
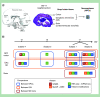
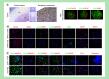
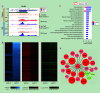
Aim: To provide a comprehensive understanding of gene regulatory networks in the developing human brain and a foundation for interpreting pathogenic deregulation. Materials & methods: We generated reference epigenomes and transcriptomes of dissected brain regions and primary neural progenitor cells (NPCs) derived from cortical and ganglionic eminence tissues of four normal human fetuses. Results: Integration of these data across developmental stages revealed a directional increase in active regulatory states, transcription factor activities and gene transcription with developmental stage. Consistent with differences in their biology, NPCs derived from cortical and ganglionic eminence regions contained common, region specific, and gestational week specific regulatory states. Conclusion: We provide a high-resolution regulatory network for NPCs from different brain regions as a comprehensive reference for future studies.
Keywords: DNA methylation; brain; cortex; enhancer; epigenetics; fetal; ganglionic eminence; gestational week; neural progenitor cells; transcriptional network.
Conflict of interest statement
This work was supported with funding provided by the US National Institutes of Health (NIH) Roadmap Epigenomics Program, NIH grant 5U01ES017154-02 and from Genome British Columbia and the Canadian Institutes of Health Research as part of the Canadian Epigenetics, Environment and Health Research Consortium Network (CIHR-262119). L Li is supported by a Genome Science and Technology Graduate Program Fellowship, University of British Columbia. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures




References
-
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17(3), 329–340 (2015). - PubMed
-
• Provides a comprehensive transcriptome mapping of neural stem cells upon injury at a single cell level.
