Influenza A virus NS1 optimises virus infectivity by enhancing genome packaging in a dsRNA-binding dependent manner
- PMID: 32677963
- PMCID: PMC7367362
- DOI: 10.1186/s12985-020-01357-3
Influenza A virus NS1 optimises virus infectivity by enhancing genome packaging in a dsRNA-binding dependent manner
Abstract
Background: The non-structural protein 1 (NS1) of influenza A virus (IAV) is a key player in inhibiting antiviral response in host cells, thereby facilitating its replication. However, other roles of NS1, which are independent of antagonising host cells' antiviral response, are less characterised.
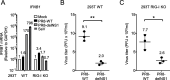
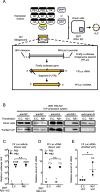
Methods: To investigate these unidentified roles, we used a recombinant virus, which lacks NS1 expression, and observed its phenotypes during the infection of antiviral defective cells (RIG-I KO cells) in the presence or absence of exogeneous NS1. Moreover, we used virus-like particle (VLP) production system to further support our findings.
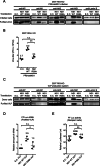
Results: Our experiments demonstrated that IAV deficient in NS1 replicates less efficiently than wild-type IAV in RIG-I KO cells and this replication defect was complemented by ectopic expression of NS1. As suggested previously, NS1 is incorporated in the virion and participates in the regulation of viral transcription and translation. Using the VLP production system, in which minigenome transcription or viral protein production was unaffected by NS1, we demonstrated that NS1 facilitates viral genome packaging into VLP, leading to efficient minigenome transfer by VLP. Furthermore, the incorporation of NS1 and the minigenome into VLP were impaired by introducing a point mutation (R38A) in the double stranded RNA-binding domain of NS1.
Conclusion: These results suggest a novel function of NS1 in improving genome packaging in a dsRNA binding-dependent manner. Taken together, NS1 acts as an essential pro-viral regulator, not only by antagonizing host immunity but also by facilitating viral replication and genome packaging.
Keywords: Genome packaging; Influenza a virus; Innate immunity; Non-structural protein 1; Virus-like production system; dsRNA binding domain.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Quantitative proteomic analysis of the influenza A virus nonstructural proteins NS1 and NS2 during natural cell infection identifies PACT as an NS1 target protein and antiviral host factor.J Virol. 2014 Aug;88(16):9038-48. doi: 10.1128/JVI.00830-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899174 Free PMC article.
-
An A14U Substitution in the 3' Noncoding Region of the M Segment of Viral RNA Supports Replication of Influenza Virus with an NS1 Deletion by Modulating Alternative Splicing of M Segment mRNAs.J Virol. 2015 Oct;89(20):10273-85. doi: 10.1128/JVI.00919-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223635 Free PMC article.
-
A Promising IFN-Deficient System to Manufacture IFN-Sensitive Influenza Vaccine Virus.Front Cell Infect Microbiol. 2018 May 1;8:127. doi: 10.3389/fcimb.2018.00127. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29765910 Free PMC article.
-
Structure and Function of the Influenza A Virus Non-Structural Protein 1.J Microbiol Biotechnol. 2019 Aug 28;29(8):1184-1192. doi: 10.4014/jmb.1903.03053. J Microbiol Biotechnol. 2019. PMID: 31154753 Review.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
Cited by
-
The C-Terminal Domains of the PB2 Subunit of the Influenza A Virus RNA Polymerase Directly Interact with Cellular GTPase Rab11a.J Virol. 2022 Mar 9;96(5):e0197921. doi: 10.1128/jvi.01979-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019720 Free PMC article.
-
How Influenza Virus Uses Host Cell Pathways during Uncoating.Cells. 2021 Jul 8;10(7):1722. doi: 10.3390/cells10071722. Cells. 2021. PMID: 34359892 Free PMC article. Review.
-
Recent Progress in Recombinant Influenza Vaccine Development Toward Heterosubtypic Immune Response.Front Immunol. 2022 May 19;13:878943. doi: 10.3389/fimmu.2022.878943. eCollection 2022. Front Immunol. 2022. PMID: 35663997 Free PMC article. Review.
References
-
- Stasakova J, Ferko B, Kittel C, Sereinig S, Romanova J, Katinger H, Egorov A. Influenza a mutant viruses with altered NS1 protein function provoke caspase-1 activation in primary human macrophages, resulting in fast apoptosis and release of high levels of interleukins 1beta and 18. J Gen Virol. 2005;86(Pt 1):185–195. doi: 10.1099/vir.0.80422-0. - DOI - PubMed
-
- Moriyama M, Chen IY, Kawaguchi A, Koshiba T, Nagata K, Takeyama H, Hasegawa H, Ichinohe T. The RNA- and TRIM25-binding domains of influenza virus NS1 protein are essential for suppression of NLRP3 Inflammasome-mediated interleukin-1beta secretion. J Virol. 2016;90(8):4105–4114. doi: 10.1128/JVI.00120-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

