Increase of vancomycin-resistant Enterococcus faecium strain type ST117 CT71 at Charité - Universitätsmedizin Berlin, 2008 to 2018
- PMID: 32678047
- PMCID: PMC7364619
- DOI: 10.1186/s13756-020-00754-1
Increase of vancomycin-resistant Enterococcus faecium strain type ST117 CT71 at Charité - Universitätsmedizin Berlin, 2008 to 2018
Abstract
Background: In addition to an overall rise in vancomycin-resistant Enterococcus faecium (VREfm), an increase in certain strain types marked by sequence type (ST) and cluster type (CT) has been reported in Germany over the past few years. Outbreak analyses at Charité - Universitätsmedizin Berlin revealed the frequent occurrence of VREfm ST117 CT71 isolates in 2017 and 2018. To investigate whether ST117 CT71 have emerged in recent years or whether these strains have been circulating for a longer time, we retrospectively analyzed non-outbreak strains that occurred between 2008 and 2018 to identify frequent STs and CTs.
Methods: In total, 120 VREfm isolates obtained from clinical and surveillance cultures from the years 2008, 2013, 2015, and 2018 were analyzed. Thirty isolates per year comprising the first 7-8 non-outbreak isolates of each quarter of the respective year were sequenced using whole genome sequencing. MLST and cgMLST were determined as well as resistance genes and virulence factors. Risk factors for VREfm ST117 were analyzed in a multivariable analysis with patient characteristics as possible confounders.
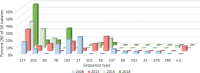
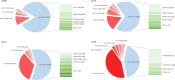
Results: The percentage of VREfm of type ST117 increased from 17% in 2008 to 57% in 2018 (p = 0.012). In 2008, vanA genotype accounted for 80% of all ST117 isolates compared to 6% in 2018. VanB CT71 first appeared in 2018 and predominated over all other ST117 at 43% (p < 0.0001). The set of resistance genes (msrC, efmA, erm(B), dfrG, aac(6')-Ii, gyrA, parC and pbp5) and virulence factors (acm, esp, hylEfm, ecbA and sgrA) in CT71 was also found in other ST117 non-CT71 strains, mainly in CT36. The study population did not differ among the different calendar years analyzed in terms of age, gender, length of stay, or ward type (each p > 0.2).
Conclusion: This study revealed an increase in ST117 strains from 2008 to 2018, accompanied by a shift toward CT71 strains with the vanB genotype in 2018. We did not detect resistance or virulence traits in CT71 that could confer survival advantage compared to other CTs among ST117 strains. To date, it is not clear why ST117 and in particular strain type ST117 CT71 predominates over other strains.
Keywords: CT71; Resistance genes; ST117; Vancomycin-resistant Enterococcus faecium; Virulence factors; Whole genome sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Chiang HY, Perencevich EN, Nair R, Nelson RE, Samore M, Khader K, et al. Incidence and outcomes associated with infections caused by Vancomycin-resistant enterococci in the United States: systematic literature review and meta-analysis. Infect Control Hosp Epidemiol. 2017;38(2):203–215. doi: 10.1017/ice.2016.254. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

