Disruption of the tumour-associated EMP3 enhances erythroid proliferation and causes the MAM-negative phenotype
- PMID: 32678083
- PMCID: PMC7366909
- DOI: 10.1038/s41467-020-17060-4
Disruption of the tumour-associated EMP3 enhances erythroid proliferation and causes the MAM-negative phenotype
Abstract
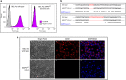
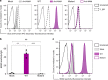
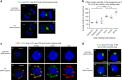
The clinically important MAM blood group antigen is present on haematopoietic cells of all humans except rare MAM-negative individuals. Its molecular basis is unknown. By whole-exome sequencing we identify EMP3, encoding epithelial membrane protein 3 (EMP3), as a candidate gene, then demonstrate inactivating mutations in ten known MAM-negative individuals. We show that EMP3, a purported tumour suppressor in various solid tumours, is expressed in erythroid cells. Disruption of EMP3 by CRISPR/Cas9 gene editing in an immortalised human erythroid cell line (BEL-A2) abolishes MAM expression. We find EMP3 to associate with, and stabilise, CD44 in the plasma membrane. Furthermore, cultured erythroid progenitor cells from MAM-negative individuals show markedly increased proliferation and higher reticulocyte yields, suggesting an important regulatory role for EMP3 in erythropoiesis and control of cell production. Our data establish MAM as a new blood group system and demonstrate an interaction of EMP3 with the cell surface signalling molecule CD44.
Conflict of interest statement
A patent application (PCT Application No. PCT/GB2019/053102) covering ex vivo erythroid cell culture proliferation enhancement due to lack of or disruption of EMP3 has been filed (N.T., C.A.G., L.T., V.K.C. and D.J.A.). The remaining authors declare no competing interests.
Figures



References
-
- Anderson G, et al. An antibody to a high frequency antigen found on red cells, platelets, lymphocytes, and monocytes. Transfusion. 1993;33:23S.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

