Single-cell RNA sequencing reveals the cellular heterogeneity of aneurysmal infrarenal abdominal aorta
- PMID: 32678909
- PMCID: PMC8064434
- DOI: 10.1093/cvr/cvaa214
Single-cell RNA sequencing reveals the cellular heterogeneity of aneurysmal infrarenal abdominal aorta
Abstract
Aims: The artery contains numerous cell types which contribute to multiple vascular diseases. However, the heterogeneity and cellular responses of these vascular cells during abdominal aortic aneurysm (AAA) progression have not been well characterized.
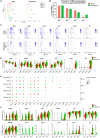
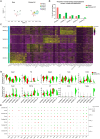
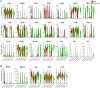
Methods and results: Single-cell RNA sequencing was performed on the infrarenal abdominal aortas (IAAs) from C57BL/6J mice at Days 7 and 14 post-sham or peri-adventitial elastase-induced AAA. Unbiased clustering analysis of the transcriptional profiles from >4500 aortic cells identified 17 clusters representing nine-cell lineages, encompassing vascular smooth muscle cells (VSMCs), fibroblasts, endothelial cells, immune cells (macrophages, T cells, B cells, and dendritic cells), and two types of rare cells, including neural cells and erythrocyte cells. Seurat clustering analysis identified four smooth muscle cell (SMC) subpopulations and five monocyte/macrophage subpopulations, with distinct transcriptional profiles. During AAA progression, three major SMC subpopulations were proportionally decreased, whereas the small subpopulation was increased, accompanied with down-regulation of SMC contractile markers and up-regulation of pro-inflammatory genes. Another AAA-associated cellular response is immune cell expansion, particularly monocytes/macrophages. Elastase exposure induced significant expansion and activation of aortic resident macrophages, blood-derived monocytes and inflammatory macrophages. We also identified increased blood-derived reparative macrophages expressing anti-inflammatory cytokines suggesting that resolution of inflammation and vascular repair also persist during AAA progression.
Conclusion: Our data identify AAA disease-relevant transcriptional signatures of vascular cells in the IAA. Furthermore, we characterize the heterogeneity and cellular responses of VSMCs and monocytes/macrophages during AAA progression, which provide insights into their function and the regulation of AAA onset and progression.
Keywords: Abdominal aortic aneurysm; Lineage heterogeneity; Macrophage; Single-cell RNA sequencing; Vascular smooth muscle cell.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author(s) 2020. For permissions, please email: journals.permissions@oup.com.
Figures




Comment in
-
Single-cell transcriptomics as a building block for determining mechanistic insight of abdominal aortic aneurysm formation.Cardiovasc Res. 2021 Apr 23;117(5):1243-1244. doi: 10.1093/cvr/cvab083. Cardiovasc Res. 2021. PMID: 33723571 Free PMC article. No abstract available.
References
-
- Nordon IM, Hinchliffe RJ, Loftus IM, Thompson MM.. Pathophysiology and epidemiology of abdominal aortic aneurysms. Nat Rev Cardiol 2011;8:92–102. - PubMed
-
- Rabkin SW. The role matrix metalloproteinases in the production of aortic aneurysm. Prog Mol Biol Transl Sci 2017;147:239–265. - PubMed
-
- Zhao G, Fu Y, Cai Z, Yu F, Gong Z, Dai R, Hu Y, Zeng L, Xu Q, Kong W.. Unspliced XBP1 confers VSMC homeostasis and prevents aortic aneurysm formation via FoxO4 interaction. Circ Res 2017;121:1331–1345. - PubMed
-
- Raffort J, Lareyre F, Clement M, Hassen-Khodja R, Chinetti G, Mallat Z.. Monocytes and macrophages in abdominal aortic aneurysm. Nat Rev Cardiol 2017;14:457–471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

