IL6/STAT3 Signaling Hijacks Estrogen Receptor α Enhancers to Drive Breast Cancer Metastasis
- PMID: 32679107
- PMCID: PMC7116707
- DOI: 10.1016/j.ccell.2020.06.007
IL6/STAT3 Signaling Hijacks Estrogen Receptor α Enhancers to Drive Breast Cancer Metastasis
Abstract
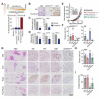
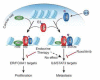
The cytokine interleukin-6 (IL6) and its downstream effector STAT3 constitute a key oncogenic pathway, which has been thought to be functionally connected to estrogen receptor α (ER) in breast cancer. We demonstrate that IL6/STAT3 signaling drives metastasis in ER+ breast cancer independent of ER. STAT3 hijacks a subset of ER enhancers to drive a distinct transcriptional program. Although these enhancers are shared by both STAT3 and ER, IL6/STAT3 activity is refractory to standard ER-targeted therapies. Instead, inhibition of STAT3 activity using the JAK inhibitor ruxolitinib decreases breast cancer invasion in vivo. Therefore, IL6/STAT3 and ER oncogenic pathways are functionally decoupled, highlighting the potential of IL6/STAT3-targeted therapies in ER+ breast cancer.
Keywords: FOXA1; IL6; STAT3; breast cancer; estrogen receptor; metastasis; mouse intraductal xenograft model; pioneer factor.
Crown Copyright © 2020. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests J.S.C. is the Founder and CSO of Azeria Therapeutics, and S.J.J. has moved to AstraZeneca during the revision of the manuscript. The other authors declare no competing interests.
Figures





References
-
- Aleskandarany MA, Agarwal D, Negm OH, Ball G, Elmouna A, Ashankyty I, Nuglozeh E, Fazaludeen MF, Diez-Rodriguez M, Nolan CC, et al. The prognostic significance of STAT3 in invasive breast cancer: analysis of protein and mRNA expressions in large cohorts. Breast Cancer Res Treat. 2016;156:9–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

