Tumor Microenvironment-Derived NRG1 Promotes Antiandrogen Resistance in Prostate Cancer
- PMID: 32679108
- PMCID: PMC7472556
- DOI: 10.1016/j.ccell.2020.06.005
Tumor Microenvironment-Derived NRG1 Promotes Antiandrogen Resistance in Prostate Cancer
Abstract
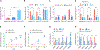
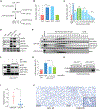
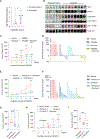
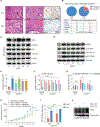
Despite the development of second-generation antiandrogens, acquired resistance to hormone therapy remains a major challenge in treating advanced prostate cancer. We find that cancer-associated fibroblasts (CAFs) can promote antiandrogen resistance in mouse models and in prostate organoid cultures. We identify neuregulin 1 (NRG1) in CAF supernatant, which promotes resistance in tumor cells through activation of HER3. Pharmacological blockade of the NRG1/HER3 axis using clinical-grade blocking antibodies re-sensitizes tumors to hormone deprivation in vitro and in vivo. Furthermore, patients with castration-resistant prostate cancer with increased tumor NRG1 activity have an inferior response to second-generation antiandrogen therapy. This work reveals a paracrine mechanism of antiandrogen resistance in prostate cancer amenable to clinical testing using available targeted therapies.
Keywords: NRG1/neuregulin 1; cancer-associated fibroblast; drug resistance; hormone therapy; tumor microenvironment.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests C.L.S. and J.W. are co-inventors of enzalutamide and apalutamide and may be entitled to royalties. C.L.S. serves on the Board of Directors of Novartis and is a co-founder of ORIC Pharmaceuticals. He is a science advisor to Agios, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ, Petra, and PMV. W.A. reports consulting for Clovis Oncology, Janssen, MORE Health, and ORIC Pharmaceuticals, and received honoraria from CARET and travel accommodations from GlaxoSmith Kline, Clovis Oncology, and ORIC Pharmaceuticals. D.E.R. reports having consulting or advisory role (uncompensated) from Genentech/Roche, Janssen Oncology, and TRACON Pharma, and received research funding from: AstraZeneca (Inst); Celgene (Inst); Ferring (Inst); Genentech/Roche (Inst); Janssen Oncology (Inst); Medivation/Astellas/Pfizer (Inst); Millennium (Inst); Novartis (Inst); Taiho Pharmaceutical (Inst); Takeda (Inst); TRACON Pharma (Inst). W.R.K. is a coinventor on patent WO2012168930A2 filed by Koninklijke Nederlandse Akademie Van Wetenschappen that covers organoid technology.
Figures




Comment in
-
Microenvironment-Mediated Resistance to Anti-Androgen Therapy.Cancer Cell. 2020 Aug 10;38(2):155-157. doi: 10.1016/j.ccell.2020.07.007. Cancer Cell. 2020. PMID: 32781041
-
NRG1 can promote antiandrogen resistance.Nat Rev Urol. 2020 Sep;17(9):486. doi: 10.1038/s41585-020-0370-4. Nat Rev Urol. 2020. PMID: 32782364 No abstract available.
References
-
- Agus DB, Sweeney CJ, Morris MJ, Mendelson DS, McNeel DG, Ahmann FR, Wang J, Derynck MK, Ng K, Lyons B, et al. (2007). Efficacy and safety of single-agent pertuzumab (rhuMAb 2C4), a human epidermal growth factor receptor dimerization inhibitor, in castration-resistant prostate cancer after progression from taxane-based therapy. J. Clin. Oncol 25, 675–681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 OD020355/OD/NIH HHS/United States
- R01 MH117406/MH/NIMH NIH HHS/United States
- R01 CA193837/CA/NCI NIH HHS/United States
- P50 CA092629/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 CA166413/CA/NCI NIH HHS/United States
- R00 CA218885/CA/NCI NIH HHS/United States
- U54 CA224079/CA/NCI NIH HHS/United States
- R01 CA204232/CA/NCI NIH HHS/United States
- R01 CA182503/CA/NCI NIH HHS/United States
- F99 CA223063/CA/NCI NIH HHS/United States
- R01 CA155169/CA/NCI NIH HHS/United States
- K99 CA218885/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

