Comparative Assessment of Protein Kinase Inhibitors in Public Databases and in PKIDB
- PMID: 32679723
- PMCID: PMC7397241
- DOI: 10.3390/molecules25143226
Comparative Assessment of Protein Kinase Inhibitors in Public Databases and in PKIDB
Abstract
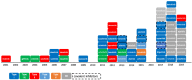
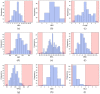
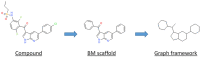
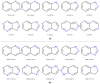
Since the first approval of a protein kinase inhibitor (PKI) by the Food and Drug Administration (FDA) in 2001, 55 new PKIs have reached the market, and many inhibitors are currently being evaluated in clinical trials. This is a clear indication that protein kinases still represent major drug targets for the pharmaceutical industry. In a previous work, we have introduced PKIDB, a publicly available database, gathering PKIs that have already been approved (Phase 4), as well as those currently in clinical trials (Phases 0 to 3). This database is updated frequently, and an analysis of the new data is presented here. In addition, we compared the set of PKIs present in PKIDB with the PKIs in early preclinical studies found in ChEMBL, the largest publicly available chemical database. For each dataset, the distribution of physicochemical descriptors related to drug-likeness is presented. From these results, updated guidelines to prioritize compounds for targeting protein kinases are proposed. The results of a principal component analysis (PCA) show that the PKIDB dataset is fully encompassed within all PKIs found in the public database. This observation is reinforced by a principal moments of inertia (PMI) analysis of all molecules. Interestingly, we notice that PKIs in clinical trials tend to explore new 3D chemical space. While a great majority of PKIs is located on the area of "flatland", we find few compounds exploring the 3D structural space. Finally, a scaffold diversity analysis of the two datasets, based on frequency counts was performed. The results give insight into the chemical space of PKIs, and can guide researchers to reach out new unexplored areas. PKIDB is freely accessible from the following website: http://www.icoa.fr/pkidb.
Keywords: approved drugs; chemometrics analysis; clinical trials; database; kinome; molecular scaffolds; protein kinase inhibitors; rings system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

