Tumors induce de novo steroid biosynthesis in T cells to evade immunity
- PMID: 32680985
- PMCID: PMC7368057
- DOI: 10.1038/s41467-020-17339-6
Tumors induce de novo steroid biosynthesis in T cells to evade immunity
Abstract
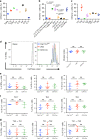
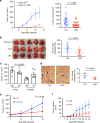
Tumors subvert immune cell function to evade immune responses, yet the complex mechanisms driving immune evasion remain poorly understood. Here we show that tumors induce de novo steroidogenesis in T lymphocytes to evade anti-tumor immunity. Using a transgenic steroidogenesis-reporter mouse line we identify and characterize de novo steroidogenic immune cells, defining the global gene expression identity of these steroid-producing immune cells and gene regulatory networks by using single-cell transcriptomics. Genetic ablation of T cell steroidogenesis restricts primary tumor growth and metastatic dissemination in mouse models. Steroidogenic T cells dysregulate anti-tumor immunity, and inhibition of the steroidogenesis pathway is sufficient to restore anti-tumor immunity. This study demonstrates T cell de novo steroidogenesis as a mechanism of anti-tumor immunosuppression and a potential druggable target.
Conflict of interest statement
In the last three years, S.A.T has consulted for Genentech and Roche, and is a member of Scientific Advisory Boards at Biogen, GlaxoSmithKline and Foresite Labs. The remaining authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

