Characterization of R-Loop Structures Using Single-Molecule R-Loop Footprinting and Sequencing
- PMID: 32681515
- PMCID: PMC7669279
- DOI: 10.1007/978-1-0716-0680-3_15
Characterization of R-Loop Structures Using Single-Molecule R-Loop Footprinting and Sequencing
Abstract
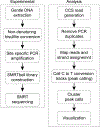
R-loops are three-stranded structures that form during transcription when the nascent RNA hybridizes with the template DNA resulting in a DNA:RNA hybrid and a looped-out single-stranded DNA (ssDNA) strand. These structures are important for normal cellular processes and aberrant R-loop formation has been implicated in a number of pathological outcomes, including certain cancers and neurodegenerative diseases. Mapping R-loops has primarily been performed using DRIP (DNA:RNA immunoprecipitation) based methods that are dependent on the anti-DNA:RNA hybrid S9.6 antibody and short-read sequencing. While DRIP-based methods are robust and report R-loop formation genome-wide, they only do so at the population average level; interrogating R-loop formation at the single molecule level is not feasible with such approaches. Here we present single molecule R-loop footprinting (SMRF-seq), a method that relies on the chemical reactivity of the displaced ssDNA strand to non-denaturing sodium bisulfite and single molecule long-read sequencing as a readout, to characterize R-loops. SMRF-seq can be used independently of S9.6 to generate high resolution, strand-specific, maps of individual R-loops at ultra-deep coverage on kilobases-length DNA fragments.
Keywords: DNA:RNA hybrid; Non-denaturing bisulfite conversion; R-loop; SMRT sequencing; Transcription.
Figures