Escherichia coli ST8196 is a novel, locally evolved, and extensively drug resistant pathogenic lineage within the ST131 clonal complex
- PMID: 32686595
- PMCID: PMC7473005
- DOI: 10.1080/22221751.2020.1797541
Escherichia coli ST8196 is a novel, locally evolved, and extensively drug resistant pathogenic lineage within the ST131 clonal complex
Abstract
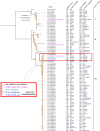
The H30Rx subclade of Escherichia coli ST131 is a clinically important, globally dispersed pathogenic lineage that typically displays resistance to fluoroquinolones and extended spectrum β-lactams. Isolates EC233 and EC234, variants of ST131-H30Rx with a novel sequence type (ST) 8196, isolated from unrelated patients presenting with bacteraemia at a Sydney Hospital in 2014 are characterised here. EC233 and EC234 are phylogroup B2, serotype O25:H4A, and resistant to ampicillin, amoxicillin, cefoxitin, ceftazidime, ceftriaxone, ciprofloxacin, norfloxacin and gentamicin and are likely clonal. Both harbour an IncFII_2 plasmid (pSPRC_Ec234-FII) that carries most of the resistance genes on an IS26 associated translocatable unit, two small plasmids and a novel IncI1 plasmid (pSPRC_Ec234-I). SNP-based phylogenetic analysis of the core genome of representatives within the ST131 clonal complex places both isolates in a subclade with three clinical Australian ST131-H30Rx clade-C isolates. A MrBayes phylogeny analysis of EC233 and EC234 indicates ST8196 share a most recent common ancestor with ST131-H30Rx strain EC70 isolated from the same hospital in 2013. Our study identified genomic hallmarks that define the ST131-H30Rx subclade in the ST8196 isolates and highlights a need for unbiased genomic surveillance approaches to identify novel high-risk MDR E. coli pathogens that impact healthcare facilities.
Keywords: E. coli ST131 clonal complex; Escherichia coli ST8196; bla CTX-M-15; qnrB4; ST131- H30Rx.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Cho S, Gupta SK, McMillan EA, et al. . Genomic analysis of multidrug-resistant Escherichia coli from surface water in Northeast Georgia, United States: presence of an ST131 epidemic strain containing blaCTX-M-15 on a phage-like plasmid. Microb Drug Resist. 2019 Nov 14;26(5):447–455. - PubMed
-
- Melo LC, Haenni M, Saras E, et al. . Emergence of the C1-M27 cluster in ST131 Escherichia coli from companion animals in France. J Antimicrob Chemother. 2019 Oct 1;74(10):3111–3113. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
