Cross-feeding modulates the rate and mechanism of antibiotic resistance evolution in a model microbial community of Escherichia coli and Salmonella enterica
- PMID: 32687537
- PMCID: PMC7392344
- DOI: 10.1371/journal.ppat.1008700
Cross-feeding modulates the rate and mechanism of antibiotic resistance evolution in a model microbial community of Escherichia coli and Salmonella enterica
Abstract
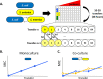
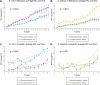
With antibiotic resistance rates on the rise, it is critical to understand how microbial species interactions influence the evolution of resistance. In obligate mutualisms, the survival of any one species (regardless of its intrinsic resistance) is contingent on the resistance of its cross-feeding partners. This sets the community antibiotic sensitivity at that of the 'weakest link' species. In this study, we tested the hypothesis that weakest link dynamics in an obligate cross-feeding relationship would limit the extent and mechanisms of antibiotic resistance evolution. We experimentally evolved an obligate co-culture and monoculture controls along gradients of two different antibiotics. We measured the rate at which each treatment increased antibiotic resistance, and sequenced terminal populations to question whether mutations differed between mono- and co-cultures. In both rifampicin and ampicillin treatments, we observed that resistance evolved more slowly in obligate co-cultures of E. coli and S. enterica than in monocultures. While we observed similar mechanisms of resistance arising under rifampicin selection, under ampicillin selection different resistance mechanisms arose in co-cultures and monocultures. In particular, mutations in an essential cell division protein, ftsI, arose in S. enterica only in co-culture. A simple mathematical model demonstrated that reliance on a partner is sufficient to slow the rate of adaptation, and can change the distribution of adaptive mutations that are acquired. Our results demonstrate that cooperative metabolic interactions can be an important modulator of resistance evolution in microbial communities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

