Phylogenetic and phylodynamic analyses of SARS-CoV-2
- PMID: 32687861
- PMCID: PMC7366979
- DOI: 10.1016/j.virusres.2020.198098
Phylogenetic and phylodynamic analyses of SARS-CoV-2
Abstract
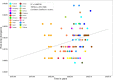
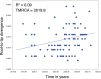
To investigate the evolutionary and epidemiological dynamics of the current COVID-19 outbreak, a total of 112 genomes of SARS-CoV-2 strains sampled from China and 12 other countries with sampling dates between 24 December 2019 and 9 February 2020 were analyzed. We performed phylogenetic, split network, likelihood-mapping, model comparison, and phylodynamic analyses of the genomes. Based on Bayesian time-scaled phylogenetic analysis with the best-fitting combination models, we estimated the time to the most recent common ancestor (TMRCA) and evolutionary rate of SARS-CoV-2 to be 12 November 2019 (95 % BCI: 11 October 2019 and 09 December 2019) and 9.90 × 10-4 substitutions per site per year (95 % BCI: 6.29 × 10-4-1.35 × 10-3), respectively. Notably, the very low Re estimates of SARS-CoV-2 during the recent sampling period may be the result of the successful control of the pandemic in China due to extreme societal lockdown efforts. Our results emphasize the importance of using phylodynamic analyses to provide insights into the roles of various interventions to limit the spread of SARS-CoV-2 in China and beyond.
Keywords: COVID-19; Evolutionary rate; Lockdown; Re; SARS-CoV-2; TMRCA.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

