Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions
- PMID: 32687918
- PMCID: PMC7367812
- DOI: 10.1016/j.lfs.2020.118102
Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions
Abstract
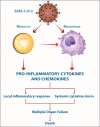
The COVID-19-, SARS- and MERS-related coronaviruses share many genomic and structural similarities. However, the SARS-CoV-2 is less pathogenic than SARS-CoV and MERS-CoV. Despite some differences in the cytokine patterns, it seems that the cytokine storm plays a crucial role in the pathogenesis of COVID-19-, SARS- and MERS. Monocytes and macrophages may be infected by SARS-CoV-2 through ACE2-dependent and ACE2-independent pathways. SARS-CoV-2 can effectively suppress the anti-viral IFN response in monocytes and macrophages. Since macrophages and dendritic cells (DCs) act as antigen presenting cells (APCs), the infection of these cells by SARS-CoV-2 impairs the adaptive immune responses against the virus. Upon infection, monocytes migrate to the tissues where they become infected resident macrophages, allowing viruses to spread through all organs and tissues. The SARS-CoV-2-infected monocytes and macrophages can produce large amounts of numerous types of pro-inflammatory cytokines and chemokines, which contribute to local tissue inflammation and a dangerous systemic inflammatory response called cytokine storm. Both local tissue inflammation and the cytokine storm play a fundamental role in the development of COVID-19-related complications, such as acute respiratory distress syndrome (ARDS), which is a main cause of death in COVID-19 patients. Here, we describe the monocytes and macrophage responses during severe coronavirus infections, while highlighting potential therapeutic interventions to attenuate macrophage-related inflammatory reactions in possible approaches for COVID-19 treatment.
Keywords: COVID-19; Macrophages; Monocytes; Pathogenesis; SARS-CoV-2.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest Authors declare that they do not have any conflicts of interest.
Figures




References
-
- Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Le infezioni in medicina. 2020;28(2):174–184. - PubMed
-
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38(1):1–9. - PubMed
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

