Quantifying epigenetic stability with minimum action paths
- PMID: 32688511
- PMCID: PMC7412882
- DOI: 10.1103/PhysRevE.101.062409
Quantifying epigenetic stability with minimum action paths
Abstract
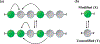
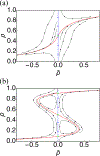
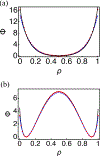
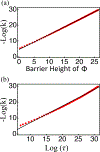
Chromatin can adopt multiple stable, heritable states with distinct histone modifications and varying levels of gene expression. Insight on the stability and maintenance of such epigenetic states can be gained by mathematical modeling of stochastic reaction networks for histone modifications. Analytical results for the kinetic networks are particularly valuable. Compared to computationally demanding numerical simulations, they often are more convenient at evaluating the robustness of conclusions with respect to model parameters. In this communication, we developed a second-quantization-based approach that can be used to analyze discrete stochastic models with a fixed, finite number of particles using a representation of the SU(2) algebra. We applied the approach to a kinetic model of chromatin states that captures the feedback between nucleosomes and the enzymes conferring histone modifications. Using a path-integral expression for the transition probability, we computed the epigenetic landscape that helps to identify the emergence of bistability and the most probable path connecting the two steady states. We anticipate the generalizability of the approach will make it useful for studying more complicated models that couple epigenetic modifications with transcription factors and chromatin structure.
Figures
Similar articles
-
Quantifying the Stability of Coupled Genetic and Epigenetic Switches With Variational Methods.Front Genet. 2021 Jan 22;11:636724. doi: 10.3389/fgene.2020.636724. eCollection 2020. Front Genet. 2021. PMID: 33552146 Free PMC article.
-
Dynamical phase transition in models that couple chromatin folding with histone modifications.Phys Rev E. 2024 May;109(5-1):054411. doi: 10.1103/PhysRevE.109.054411. Phys Rev E. 2024. PMID: 38907407
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Chromatin profiling and state predictions reveal insights into epigenetic regulation during early porcine development.Epigenetics Chromatin. 2024 May 21;17(1):16. doi: 10.1186/s13072-024-00542-w. Epigenetics Chromatin. 2024. PMID: 38773546 Free PMC article.
-
Interplay between different epigenetic modifications and mechanisms.Adv Genet. 2010;70:101-41. doi: 10.1016/B978-0-12-380866-0.60005-8. Adv Genet. 2010. PMID: 20920747 Review.
Cited by
-
Quantifying the Stability of Coupled Genetic and Epigenetic Switches With Variational Methods.Front Genet. 2021 Jan 22;11:636724. doi: 10.3389/fgene.2020.636724. eCollection 2020. Front Genet. 2021. PMID: 33552146 Free PMC article.
-
Preserving condensate structure and composition by lowering sequence complexity.Biophys J. 2024 Jul 2;123(13):1815-1826. doi: 10.1016/j.bpj.2024.05.026. Epub 2024 May 31. Biophys J. 2024. PMID: 38824391 Free PMC article.
-
Characterizing chromatin folding coordinate and landscape with deep learning.PLoS Comput Biol. 2020 Sep 28;16(9):e1008262. doi: 10.1371/journal.pcbi.1008262. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32986691 Free PMC article.
-
A Dimension Reduction Approach for Energy Landscape: Identifying Intermediate States in Metabolism-EMT Network.Adv Sci (Weinh). 2021 Mar 18;8(10):2003133. doi: 10.1002/advs.202003133. eCollection 2021 May. Adv Sci (Weinh). 2021. PMID: 34026435 Free PMC article.
-
Multiscale modeling of genome organization with maximum entropy optimization.J Chem Phys. 2021 Jul 7;155(1):010901. doi: 10.1063/5.0044150. J Chem Phys. 2021. PMID: 34241389 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
