Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication
- PMID: 32690711
- PMCID: PMC7414174
- DOI: 10.1073/pnas.2006165117
Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication
Abstract
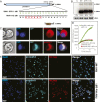
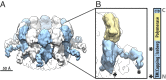
For positive-strand RNA [(+)RNA] viruses, the major target for antiviral therapies is genomic RNA replication, which occurs at poorly understood membrane-bound viral RNA replication complexes. Recent cryoelectron microscopy (cryo-EM) of nodavirus RNA replication complexes revealed that the viral double-stranded RNA replication template is coiled inside a 30- to 90-nm invagination of the outer mitochondrial membrane, whose necked aperture to the cytoplasm is gated by a 12-fold symmetric, 35-nm diameter "crown" complex that contains multifunctional viral RNA replication protein A. Here we report optimizing cryo-EM tomography and image processing to improve crown resolution from 33 to 8.5 Å. This resolves the crown into 12 distinct vertical segments, each with 3 major subdomains: A membrane-connected basal lobe and an apical lobe that together comprise the ∼19-nm-diameter central turret, and a leg emerging from the basal lobe that connects to the membrane at ∼35-nm diameter. Despite widely varying replication vesicle diameters, the resulting two rings of membrane interaction sites constrain the vesicle neck to a highly uniform shape. Labeling protein A with a His-tag that binds 5-nm Ni-nanogold allowed cryo-EM tomography mapping of the C terminus of protein A to the apical lobe, which correlates well with the predicted structure of the C-proximal polymerase domain of protein A. These and other results indicate that the crown contains 12 copies of protein A arranged basally to apically in an N-to-C orientation. Moreover, the apical polymerase localization has significant mechanistic implications for template RNA recruitment and (-) and (+)RNA synthesis.
Keywords: cryotomography; nodavirus; positive-strand RNA virus; replication complexes; replication crown.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
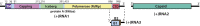





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

