Single-molecule visualization of DNA G-quadruplex formation in live cells
- PMID: 32690897
- PMCID: PMC7610488
- DOI: 10.1038/s41557-020-0506-4
Single-molecule visualization of DNA G-quadruplex formation in live cells
Abstract
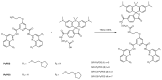
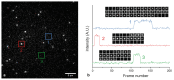
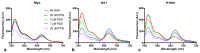
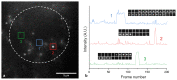
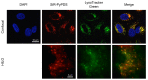
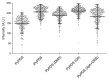
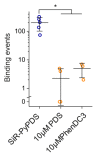
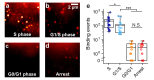
Substantial evidence now exists to support that formation of DNA G-quadruplexes (G4s) is coupled to altered gene expression. However, approaches that allow us to probe G4s in living cells without perturbing their folding dynamics are required to understand their biological roles in greater detail. Herein, we report a G4-specific fluorescent probe (SiR-PyPDS) that enables single-molecule and real-time detection of individual G4 structures in living cells. Live-cell single-molecule fluorescence imaging of G4s was carried out under conditions that use low concentrations of SiR-PyPDS (20 nM) to provide informative measurements representative of the population of G4s in living cells, without globally perturbing G4 formation and dynamics. Single-molecule fluorescence imaging and time-dependent chemical trapping of unfolded G4s in living cells reveal that G4s fluctuate between folded and unfolded states. We also demonstrate that G4 formation in live cells is cell-cycle-dependent and disrupted by chemical inhibition of transcription and replication. Our observations provide robust evidence in support of dynamic G4 formation in living cells.
Conflict of interest statement
S.B. is a founder and shareholder of Cambridge Epigenetix Ltd.
Figures






References
-
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
-
- Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol. 2017;18:279–284. - PubMed
-
- Chambers, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877–881. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

