Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming
- PMID: 32690944
- PMCID: PMC7502540
- DOI: 10.1038/s41589-020-0584-z
Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming
Abstract
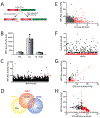
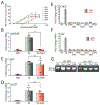
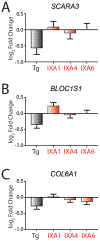
Activation of the IRE1/XBP1s signaling arm of the unfolded protein response (UPR) is a promising strategy to correct defects in endoplasmic reticulum (ER) proteostasis implicated in diverse diseases. However, no pharmacologic activators of this pathway identified to date are suitable for ER proteostasis remodeling through selective activation of IRE1/XBP1s signaling. Here, we use high-throughput screening to identify non-toxic compounds that induce ER proteostasis remodeling through IRE1/XBP1s activation. We employ transcriptional profiling to stringently confirm that our prioritized compounds selectively activate IRE1/XBP1s signaling without activating other cellular stress-responsive signaling pathways. Furthermore, we demonstrate that our compounds improve ER proteostasis of destabilized variants of amyloid precursor protein (APP) through an IRE1-dependent mechanism and reduce APP-associated mitochondrial toxicity in cellular models. These results establish highly selective IRE1/XBP1s activating compounds that can be widely employed to define the functional importance of IRE1/XBP1s activity for ER proteostasis regulation in the context of health and disease.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
Jeffrey W. Kelly is a co-founder of Proteostasis Therapeutics Inc. A patent has been submitted for compounds identified through work described in this manuscript. This patent includes R. Luke Wiseman, Jeffery W. Kelly, Julia M.D. Grandjean, Lars Plate, and Christina B. Cooley as co-inventors. No other author has a conflict of interest.
Figures
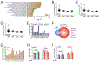

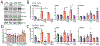
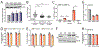

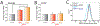
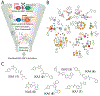
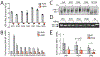
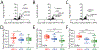

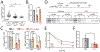
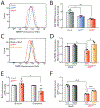
Comment in
-
A stress-free stress response.Nat Chem Biol. 2020 Oct;16(10):1038-1039. doi: 10.1038/s41589-020-0616-8. Nat Chem Biol. 2020. PMID: 32704180 No abstract available.
References
-
- Walter P; Ron D, The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011, 334 (6059), 1081–6. - PubMed
-
- Bernales S; Papa FR; Walter P, Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 2006, 22, 487–508. - PubMed
-
- Schroder M; Kaufman RJ, ER stress and the unfolded protein response. Mutat Res 2005, 569 (1–2), 29–63. - PubMed
-
- Patil C; Walter P, Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 2001, 13 (3), 349–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

